Gene
KWMTBOMO06619
Pre Gene Modal
BGIBMGA011687
Annotation
PREDICTED:_leukocyte_receptor_cluster_member_8_homolog_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.737
Sequence
CDS
ATGAAAAGTGTAAGAGAATTGGGATATAGAACCAATACCAAGTGTCCACAGTCAAAATCTAGGAGTCCTAGAAAAAGGTTAAGTGCGTCATCTTGTTCGAGTGATGAAATTGAAGAGAAAAAACCTCCAAAAGGTAAGGGGAGGCAAAAAGTTAAGGATAGGTTGTCGTTAGTTTCAAATAAAAAATCAGAAAAATTTAGCAAAAATAAAAAGAAACAGTGTAATCAGTTTGTTGTTGAAGATGCTCAGGGTAATGCTGAAAAACTTCAAAAAAGAGCTGAAAGATTTGCAGGTTCAAGTAATGTACCCTCCATAGCAAGTTCACTACAAGTAAATAGCAAAAGAGTAGAACTTACTTCTAAAAAATCAATAATCCAAGACAATGAAGGTGATTATGAAGTAAATGATATGCACATAGTGGGTACATGTCTAGATATAGAAAAGTCATTCTTGAGACTAACTAAAGCACCTGAAGCATGTGAAGTTCGCCCTGTGTCAGTTTTAAATAAGTCCCTCAAGAATGTCAAAGAAAAATGGATAGAGCGCCAAGACTATAGATATGCTTGTGATCAACTCAAATCCATCAGGCAAGATCTAACTGTGCAAGGGGTAAGAGATAGTTTCACTGTAGAAGTATATGAAACACATGCTCGCATTGCCTTGGAAAAAGGTGACCATGAGGAATTTAATCAGTGTCAAACTCAACTTAAAATGTTGTACAGTGAATTGCCACACTGTAGATCAAATTTAGCAGAATTTACAGCATACAGGATATTGTACTACATTTTTACAAAGAATACATTAGATTTAACAACCATATTTCAGTTTCTATCTAAAGAAGACAGGGAAAATAATTGTATAAAACATGCATTACAAACTAGATGTGCCTGGGCAACTGGAAACTTACACAAGTTTTTCTTACTCTACAAAACTGCCCCACTAATGGCTGGATACCTCATGGACTGGTTTGTGGAGAGGGAGCGAAAACAATACCTCAAATACATCATAAAGTCCTACAGACAGACCGTTCCTGTGGAATTCATCGCTCGAGAGTTGGCATTCGAGTCGAGTTCTAAGGCACTTGAATTTTTAAACCAATTTCCTCTATCATATGTTGGTTGTAATCGGACACAAATAGATTGCAAAACTAGCGCCACTGCTGTTACAATTTTATCAGCTTCATTAGGAGGTGGCACCAGATGA
Protein
MKSVRELGYRTNTKCPQSKSRSPRKRLSASSCSSDEIEEKKPPKGKGRQKVKDRLSLVSNKKSEKFSKNKKKQCNQFVVEDAQGNAEKLQKRAERFAGSSNVPSIASSLQVNSKRVELTSKKSIIQDNEGDYEVNDMHIVGTCLDIEKSFLRLTKAPEACEVRPVSVLNKSLKNVKEKWIERQDYRYACDQLKSIRQDLTVQGVRDSFTVEVYETHARIALEKGDHEEFNQCQTQLKMLYSELPHCRSNLAEFTAYRILYYIFTKNTLDLTTIFQFLSKEDRENNCIKHALQTRCAWATGNLHKFFLLYKTAPLMAGYLMDWFVERERKQYLKYIIKSYRQTVPVEFIARELAFESSSKALEFLNQFPLSYVGCNRTQIDCKTSATAVTILSASLGGGTR
Summary
Uniprot
A0A2A4JVH2
H9JQ81
A0A0L7LLQ5
A0A194PVK7
A0A2H1X140
S4PGX6
+ More
A0A194QNU5 A0A212FJG9 A0A2A4JTS1 A0A0T6AX67 A0A1Y1LB39 A0A1W4XLA3 A0A1Y1LIM1 A0A067QKB3 A0A1L8DGR5 T1PIJ5 A0A1B6GA47 A0A1Y1LDB6 A0A0L0C1R9 A0A1Y1LB86 F4WV85 A0A1L8DHB9 A0A1L8DGG0 A0A2J7RER9 E2AEL1 A0A195EVY8 A0A158NNW6 A0A195BRV5 A0A151WJV2 A0A1I8MDW2 A0A151IX11 N6TV86 K7ITA9 A0A0M4E968 A0A151I986 A0A1A9WUY0 A0A154PQS7 A0A0Q5WMK6 Q171R8 A0A087ZVF9 V9IAM3 A0A026WRN3 A0A2A3E7C2 B3N526 A0A1A9UQZ3 A0A1I8P6R7 A0A310SJY4 A0A0L7QS83 E2C7R6 A0A1B0FH49 A0A1Q3FIX1 B4P124 A0A0M8ZWR4 A0A1S4FH03 A0A0J9R0E3 A0A1B0A3V9 B4Q9U5 A0A182GMS1 A0A1A9XVS8 A0A1B0ANW0 A0A1W4W5L6 Q9VKP5 B4GT08 B0WRQ8 B5DJP1 A0A139WLF6 B3MJW6 A0A182TBD2 A0A3B0K9N7 A0A182P8A5 B3DNL8 W8B6N1 A0A1B6DZI1 A0A182MN61 A0A1S4GZK4 A0A182I7A2 A0A182LMU7 A0A182VUD1 B4MU50 A0A2M3YYG6 E9IH97 A0A0K8VVY1 A0A182RSJ1 A0A182YRP3 W5JT08 A0A1Y1LB83 A0A182MZ81 A0A182JZA3 B4KKM6 A0A1J1HLD2 A0A1Y1LFY0 A0A182V4A5 B4JPU5 A0A0A1X7N0 A0A182XNP3 A0A2C9M9L4 A0A182JIG0
A0A194QNU5 A0A212FJG9 A0A2A4JTS1 A0A0T6AX67 A0A1Y1LB39 A0A1W4XLA3 A0A1Y1LIM1 A0A067QKB3 A0A1L8DGR5 T1PIJ5 A0A1B6GA47 A0A1Y1LDB6 A0A0L0C1R9 A0A1Y1LB86 F4WV85 A0A1L8DHB9 A0A1L8DGG0 A0A2J7RER9 E2AEL1 A0A195EVY8 A0A158NNW6 A0A195BRV5 A0A151WJV2 A0A1I8MDW2 A0A151IX11 N6TV86 K7ITA9 A0A0M4E968 A0A151I986 A0A1A9WUY0 A0A154PQS7 A0A0Q5WMK6 Q171R8 A0A087ZVF9 V9IAM3 A0A026WRN3 A0A2A3E7C2 B3N526 A0A1A9UQZ3 A0A1I8P6R7 A0A310SJY4 A0A0L7QS83 E2C7R6 A0A1B0FH49 A0A1Q3FIX1 B4P124 A0A0M8ZWR4 A0A1S4FH03 A0A0J9R0E3 A0A1B0A3V9 B4Q9U5 A0A182GMS1 A0A1A9XVS8 A0A1B0ANW0 A0A1W4W5L6 Q9VKP5 B4GT08 B0WRQ8 B5DJP1 A0A139WLF6 B3MJW6 A0A182TBD2 A0A3B0K9N7 A0A182P8A5 B3DNL8 W8B6N1 A0A1B6DZI1 A0A182MN61 A0A1S4GZK4 A0A182I7A2 A0A182LMU7 A0A182VUD1 B4MU50 A0A2M3YYG6 E9IH97 A0A0K8VVY1 A0A182RSJ1 A0A182YRP3 W5JT08 A0A1Y1LB83 A0A182MZ81 A0A182JZA3 B4KKM6 A0A1J1HLD2 A0A1Y1LFY0 A0A182V4A5 B4JPU5 A0A0A1X7N0 A0A182XNP3 A0A2C9M9L4 A0A182JIG0
Pubmed
19121390
26227816
26354079
23622113
22118469
28004739
+ More
24845553 26108605 21719571 20798317 21347285 25315136 23537049 20075255 17994087 17510324 24508170 17550304 22936249 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18362917 19820115 24495485 12364791 20966253 21282665 25244985 20920257 23761445 25830018 15562597
24845553 26108605 21719571 20798317 21347285 25315136 23537049 20075255 17994087 17510324 24508170 17550304 22936249 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18362917 19820115 24495485 12364791 20966253 21282665 25244985 20920257 23761445 25830018 15562597
EMBL
NWSH01000601
PCG75410.1
BABH01012019
JTDY01000642
KOB76360.1
KQ459590
+ More
KPI97357.1 ODYU01012538 SOQ58916.1 GAIX01003467 JAA89093.1 KQ461190 KPJ07187.1 AGBW02008266 OWR53888.1 PCG75411.1 LJIG01022596 KRT79740.1 GEZM01060822 JAV70879.1 GEZM01060824 JAV70877.1 KK853239 KDR09498.1 GFDF01008436 JAV05648.1 KA647753 AFP62382.1 GECZ01017778 GECZ01010606 JAS51991.1 JAS59163.1 GEZM01060823 JAV70878.1 JRES01001119 KNC25359.1 GEZM01060819 JAV70883.1 GL888384 EGI61851.1 GFDF01008309 JAV05775.1 GFDF01008548 JAV05536.1 NEVH01004957 PNF39331.1 GL438870 EFN68134.1 KQ981953 KYN32430.1 ADTU01021859 KQ976417 KYM89762.1 KQ983031 KYQ48138.1 KQ980843 KYN12313.1 APGK01050935 KB741178 KB632325 ENN73185.1 ERL92375.1 AAZX01003326 CP012523 ALC38688.1 KQ978303 KYM95334.1 KQ435007 KZC13618.1 CH954177 KQS70591.1 CH477447 EAT40749.1 JR038238 AEY58173.1 KK107128 EZA58351.1 KZ288345 PBC27647.1 EDV58971.1 KQ762207 OAD56043.1 KQ414766 KOC61399.1 GL453395 EFN76049.1 CCAG010021908 GFDL01007570 JAV27475.1 CM000157 EDW87999.1 KQ435845 KOX71183.1 CM002910 KMY89628.1 CM000361 EDX04612.1 JXUM01075043 KQ562865 KXJ74975.1 JXJN01001068 AE014134 AAF53017.1 ACZ94231.1 CH479189 EDW25517.1 DS232058 EDS33462.1 CH379061 EDY70513.1 KQ971321 KYB28780.1 CH902620 EDV32421.1 OUUW01000006 SPP81741.1 BT033006 ACD99570.1 GAMC01012283 JAB94272.1 GEDC01006234 JAS31064.1 AXCM01000345 AAAB01008984 APCN01003374 CH963852 EDW75639.2 GGFM01000539 MBW21290.1 GL763196 EFZ20045.1 GDHF01009276 JAI43038.1 ADMH02000239 ETN67276.1 GEZM01060818 JAV70884.1 CH933807 EDW12690.1 CVRI01000010 CRK88837.1 GEZM01060825 JAV70875.1 CH916372 EDV98925.1 GBXI01007406 JAD06886.1
KPI97357.1 ODYU01012538 SOQ58916.1 GAIX01003467 JAA89093.1 KQ461190 KPJ07187.1 AGBW02008266 OWR53888.1 PCG75411.1 LJIG01022596 KRT79740.1 GEZM01060822 JAV70879.1 GEZM01060824 JAV70877.1 KK853239 KDR09498.1 GFDF01008436 JAV05648.1 KA647753 AFP62382.1 GECZ01017778 GECZ01010606 JAS51991.1 JAS59163.1 GEZM01060823 JAV70878.1 JRES01001119 KNC25359.1 GEZM01060819 JAV70883.1 GL888384 EGI61851.1 GFDF01008309 JAV05775.1 GFDF01008548 JAV05536.1 NEVH01004957 PNF39331.1 GL438870 EFN68134.1 KQ981953 KYN32430.1 ADTU01021859 KQ976417 KYM89762.1 KQ983031 KYQ48138.1 KQ980843 KYN12313.1 APGK01050935 KB741178 KB632325 ENN73185.1 ERL92375.1 AAZX01003326 CP012523 ALC38688.1 KQ978303 KYM95334.1 KQ435007 KZC13618.1 CH954177 KQS70591.1 CH477447 EAT40749.1 JR038238 AEY58173.1 KK107128 EZA58351.1 KZ288345 PBC27647.1 EDV58971.1 KQ762207 OAD56043.1 KQ414766 KOC61399.1 GL453395 EFN76049.1 CCAG010021908 GFDL01007570 JAV27475.1 CM000157 EDW87999.1 KQ435845 KOX71183.1 CM002910 KMY89628.1 CM000361 EDX04612.1 JXUM01075043 KQ562865 KXJ74975.1 JXJN01001068 AE014134 AAF53017.1 ACZ94231.1 CH479189 EDW25517.1 DS232058 EDS33462.1 CH379061 EDY70513.1 KQ971321 KYB28780.1 CH902620 EDV32421.1 OUUW01000006 SPP81741.1 BT033006 ACD99570.1 GAMC01012283 JAB94272.1 GEDC01006234 JAS31064.1 AXCM01000345 AAAB01008984 APCN01003374 CH963852 EDW75639.2 GGFM01000539 MBW21290.1 GL763196 EFZ20045.1 GDHF01009276 JAI43038.1 ADMH02000239 ETN67276.1 GEZM01060818 JAV70884.1 CH933807 EDW12690.1 CVRI01000010 CRK88837.1 GEZM01060825 JAV70875.1 CH916372 EDV98925.1 GBXI01007406 JAD06886.1
Proteomes
UP000218220
UP000005204
UP000037510
UP000053268
UP000053240
UP000007151
+ More
UP000192223 UP000027135 UP000037069 UP000007755 UP000235965 UP000000311 UP000078541 UP000005205 UP000078540 UP000075809 UP000095301 UP000078492 UP000019118 UP000030742 UP000002358 UP000092553 UP000078542 UP000091820 UP000076502 UP000008711 UP000008820 UP000005203 UP000053097 UP000242457 UP000078200 UP000095300 UP000053825 UP000008237 UP000092444 UP000002282 UP000053105 UP000092445 UP000000304 UP000069940 UP000249989 UP000092443 UP000092460 UP000192221 UP000000803 UP000008744 UP000002320 UP000001819 UP000007266 UP000007801 UP000075901 UP000268350 UP000075885 UP000075883 UP000075840 UP000075882 UP000075920 UP000007798 UP000075900 UP000076408 UP000000673 UP000075884 UP000075881 UP000009192 UP000183832 UP000075903 UP000001070 UP000076407 UP000076420 UP000075880
UP000192223 UP000027135 UP000037069 UP000007755 UP000235965 UP000000311 UP000078541 UP000005205 UP000078540 UP000075809 UP000095301 UP000078492 UP000019118 UP000030742 UP000002358 UP000092553 UP000078542 UP000091820 UP000076502 UP000008711 UP000008820 UP000005203 UP000053097 UP000242457 UP000078200 UP000095300 UP000053825 UP000008237 UP000092444 UP000002282 UP000053105 UP000092445 UP000000304 UP000069940 UP000249989 UP000092443 UP000092460 UP000192221 UP000000803 UP000008744 UP000002320 UP000001819 UP000007266 UP000007801 UP000075901 UP000268350 UP000075885 UP000075883 UP000075840 UP000075882 UP000075920 UP000007798 UP000075900 UP000076408 UP000000673 UP000075884 UP000075881 UP000009192 UP000183832 UP000075903 UP000001070 UP000076407 UP000076420 UP000075880
Pfam
PF03399 SAC3_GANP
ProteinModelPortal
A0A2A4JVH2
H9JQ81
A0A0L7LLQ5
A0A194PVK7
A0A2H1X140
S4PGX6
+ More
A0A194QNU5 A0A212FJG9 A0A2A4JTS1 A0A0T6AX67 A0A1Y1LB39 A0A1W4XLA3 A0A1Y1LIM1 A0A067QKB3 A0A1L8DGR5 T1PIJ5 A0A1B6GA47 A0A1Y1LDB6 A0A0L0C1R9 A0A1Y1LB86 F4WV85 A0A1L8DHB9 A0A1L8DGG0 A0A2J7RER9 E2AEL1 A0A195EVY8 A0A158NNW6 A0A195BRV5 A0A151WJV2 A0A1I8MDW2 A0A151IX11 N6TV86 K7ITA9 A0A0M4E968 A0A151I986 A0A1A9WUY0 A0A154PQS7 A0A0Q5WMK6 Q171R8 A0A087ZVF9 V9IAM3 A0A026WRN3 A0A2A3E7C2 B3N526 A0A1A9UQZ3 A0A1I8P6R7 A0A310SJY4 A0A0L7QS83 E2C7R6 A0A1B0FH49 A0A1Q3FIX1 B4P124 A0A0M8ZWR4 A0A1S4FH03 A0A0J9R0E3 A0A1B0A3V9 B4Q9U5 A0A182GMS1 A0A1A9XVS8 A0A1B0ANW0 A0A1W4W5L6 Q9VKP5 B4GT08 B0WRQ8 B5DJP1 A0A139WLF6 B3MJW6 A0A182TBD2 A0A3B0K9N7 A0A182P8A5 B3DNL8 W8B6N1 A0A1B6DZI1 A0A182MN61 A0A1S4GZK4 A0A182I7A2 A0A182LMU7 A0A182VUD1 B4MU50 A0A2M3YYG6 E9IH97 A0A0K8VVY1 A0A182RSJ1 A0A182YRP3 W5JT08 A0A1Y1LB83 A0A182MZ81 A0A182JZA3 B4KKM6 A0A1J1HLD2 A0A1Y1LFY0 A0A182V4A5 B4JPU5 A0A0A1X7N0 A0A182XNP3 A0A2C9M9L4 A0A182JIG0
A0A194QNU5 A0A212FJG9 A0A2A4JTS1 A0A0T6AX67 A0A1Y1LB39 A0A1W4XLA3 A0A1Y1LIM1 A0A067QKB3 A0A1L8DGR5 T1PIJ5 A0A1B6GA47 A0A1Y1LDB6 A0A0L0C1R9 A0A1Y1LB86 F4WV85 A0A1L8DHB9 A0A1L8DGG0 A0A2J7RER9 E2AEL1 A0A195EVY8 A0A158NNW6 A0A195BRV5 A0A151WJV2 A0A1I8MDW2 A0A151IX11 N6TV86 K7ITA9 A0A0M4E968 A0A151I986 A0A1A9WUY0 A0A154PQS7 A0A0Q5WMK6 Q171R8 A0A087ZVF9 V9IAM3 A0A026WRN3 A0A2A3E7C2 B3N526 A0A1A9UQZ3 A0A1I8P6R7 A0A310SJY4 A0A0L7QS83 E2C7R6 A0A1B0FH49 A0A1Q3FIX1 B4P124 A0A0M8ZWR4 A0A1S4FH03 A0A0J9R0E3 A0A1B0A3V9 B4Q9U5 A0A182GMS1 A0A1A9XVS8 A0A1B0ANW0 A0A1W4W5L6 Q9VKP5 B4GT08 B0WRQ8 B5DJP1 A0A139WLF6 B3MJW6 A0A182TBD2 A0A3B0K9N7 A0A182P8A5 B3DNL8 W8B6N1 A0A1B6DZI1 A0A182MN61 A0A1S4GZK4 A0A182I7A2 A0A182LMU7 A0A182VUD1 B4MU50 A0A2M3YYG6 E9IH97 A0A0K8VVY1 A0A182RSJ1 A0A182YRP3 W5JT08 A0A1Y1LB83 A0A182MZ81 A0A182JZA3 B4KKM6 A0A1J1HLD2 A0A1Y1LFY0 A0A182V4A5 B4JPU5 A0A0A1X7N0 A0A182XNP3 A0A2C9M9L4 A0A182JIG0
PDB
3T5V
E-value=0.0368228,
Score=87
Ontologies
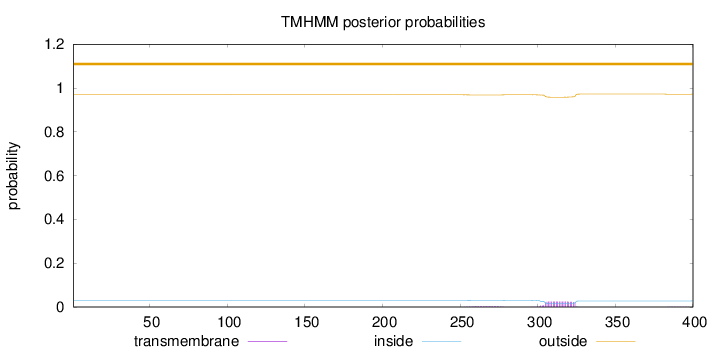
Topology
Length:
400
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.572
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03094
outside
1 - 400
Population Genetic Test Statistics
Pi
182.564249
Theta
189.59407
Tajima's D
-0.338233
CLR
0.368403
CSRT
0.28033598320084
Interpretation
Uncertain
