Gene
KWMTBOMO06618
Pre Gene Modal
BGIBMGA011686
Annotation
PREDICTED:_uncharacterized_protein_C16orf52_homolog_A_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.972
Sequence
CDS
ATGGATAAGTTAACAGTGATATCCGCCACACTTTTCATGGCTGCTGATGTTTTTGCCATTGTAAGTCTTGCAATGCCTGATTGGATAATTACAGACATTGGAGGTGACACTCGTCTAGGTCTTATGTGGTCTTGCATGACTCTATACAACAGACCTCAAGTTTGTTTTACTCCAGACCTTCAACCTGAATGGCTTTTGGCACTTATTTGCATTTTCATTGGCTGCATATGCATCACCACCACTGTTATACTATTAGCTTCAAGTCATTTTGACAGAAATGTGGTCCCATATGCAAGATGGGTTGGTTTTGCAGCAATGGTTGTATTCTGCTTAGCGGCAGTCATATTTCCCATGGGTTTCCACGTGGATGAGATTGGAGGACAACCTTATCAATTACCAAACAGTCATCAAGTTGGAATCTCCTACATATTATTTGTTCTCTCCCTTTGGATAACAGTGATATCTGAACTTTTTGCTGGGAAAGTGTGTCTCCCACACTTTTAA
Protein
MDKLTVISATLFMAADVFAIVSLAMPDWIITDIGGDTRLGLMWSCMTLYNRPQVCFTPDLQPEWLLALICIFIGCICITTTVILLASSHFDRNVVPYARWVGFAAMVVFCLAAVIFPMGFHVDEIGGQPYQLPNSHQVGISYILFVLSLWITVISELFAGKVCLPHF
Summary
Uniprot
H9JQ80
S4NW74
A0A194PVD4
A0A2H1X0P3
A0A2A4JUN5
A0A1Y1NKJ7
+ More
A0A1B6L7W2 A0A2P8XKX0 A0A1B6EGH6 A0A1W4X8Z3 A0A067QMS5 A0A0L7RIV7 A0A088AHU8 D6WF05 A0A194QNT6 A0A232EZV7 K7IYN0 R4WDU3 A0A2R7VVG0 A0A2A3EA52 A0A0C9RUD9 A0A1L8D9N0 A0A1B0EZ85 A0A2H8TZP6 A0A2S2QLD0 R4G8K6 J9K0V3 A0A224XRA5 A0A1S3DK63 A0A154PS35 A0A1Q3FM68 A0A182QK32 B0WDU2 Q17FB9 A0A084WHG7 J3JXW7 A0A1I8M150 A0A0L0CEC6 A0A2C9H8P1 A0A182Y207 A0A2C9GQU6 A0A182SA86 A0A182UM93 A0A182U4L7 A0A182KSX6 Q7Q020 B4LBK6 B4L0V3 A0A1I8PBL6 B4IZL5 A0A182JXQ0 A0A023EHT8 A0A182MCP6 A0A1B0FE16 A0A1A9Y2S1 A0A1A9VKV4 A0A1B0AWC7 A0A182WR14 A0A1B0A687 A0A1A9WSS8 A0A0N7Z920 A0A182P1R3 A0A2M4AXC0 A0A2M4C1I8 W5JIF5 A0A0K8TKB7 E2A9K9 B4MMI6 A0A3B0JV77 Q2LYW3 B4H024 W8BFY0 B3M8S3 A0A0M4F0H8 A0A1W4W6U6 E9J5Y1 B4IIT3 B3NE14 B4QRD9 Q9VW61 B4PFV3 T1GL01 A0A026WTR4 A0A151K4E5 A0A195E808 F4X6K5 A0A151I176 A0A151XBD5 A0A336MSD7 A0A182RQV3 A0A158NGF7 A0A0P5KGR4 E9FW33 A0A3L8DHP5 A0A151ID99 A0A087TBE3 A0A0N8DNX8 U5EVX3 A0A182N2M3 T1J1T7
A0A1B6L7W2 A0A2P8XKX0 A0A1B6EGH6 A0A1W4X8Z3 A0A067QMS5 A0A0L7RIV7 A0A088AHU8 D6WF05 A0A194QNT6 A0A232EZV7 K7IYN0 R4WDU3 A0A2R7VVG0 A0A2A3EA52 A0A0C9RUD9 A0A1L8D9N0 A0A1B0EZ85 A0A2H8TZP6 A0A2S2QLD0 R4G8K6 J9K0V3 A0A224XRA5 A0A1S3DK63 A0A154PS35 A0A1Q3FM68 A0A182QK32 B0WDU2 Q17FB9 A0A084WHG7 J3JXW7 A0A1I8M150 A0A0L0CEC6 A0A2C9H8P1 A0A182Y207 A0A2C9GQU6 A0A182SA86 A0A182UM93 A0A182U4L7 A0A182KSX6 Q7Q020 B4LBK6 B4L0V3 A0A1I8PBL6 B4IZL5 A0A182JXQ0 A0A023EHT8 A0A182MCP6 A0A1B0FE16 A0A1A9Y2S1 A0A1A9VKV4 A0A1B0AWC7 A0A182WR14 A0A1B0A687 A0A1A9WSS8 A0A0N7Z920 A0A182P1R3 A0A2M4AXC0 A0A2M4C1I8 W5JIF5 A0A0K8TKB7 E2A9K9 B4MMI6 A0A3B0JV77 Q2LYW3 B4H024 W8BFY0 B3M8S3 A0A0M4F0H8 A0A1W4W6U6 E9J5Y1 B4IIT3 B3NE14 B4QRD9 Q9VW61 B4PFV3 T1GL01 A0A026WTR4 A0A151K4E5 A0A195E808 F4X6K5 A0A151I176 A0A151XBD5 A0A336MSD7 A0A182RQV3 A0A158NGF7 A0A0P5KGR4 E9FW33 A0A3L8DHP5 A0A151ID99 A0A087TBE3 A0A0N8DNX8 U5EVX3 A0A182N2M3 T1J1T7
Pubmed
19121390
23622113
26354079
28004739
29403074
24845553
+ More
18362917 19820115 28648823 20075255 23691247 17510324 24438588 22516182 23537049 25315136 26108605 25244985 20966253 12364791 14747013 17210077 17994087 24945155 26483478 27129103 20920257 23761445 26369729 20798317 15632085 24495485 21282665 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24508170 21719571 21347285 21292972 30249741
18362917 19820115 28648823 20075255 23691247 17510324 24438588 22516182 23537049 25315136 26108605 25244985 20966253 12364791 14747013 17210077 17994087 24945155 26483478 27129103 20920257 23761445 26369729 20798317 15632085 24495485 21282665 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24508170 21719571 21347285 21292972 30249741
EMBL
BABH01012019
GAIX01009544
JAA83016.1
KQ459590
KPI97356.1
ODYU01012538
+ More
SOQ58915.1 NWSH01000601 PCG75408.1 GEZM01000267 JAV98444.1 GEBQ01020180 JAT19797.1 PYGN01001825 PSN32649.1 GEDC01000279 JAS37019.1 KK853239 KDR09495.1 KQ414583 KOC70743.1 KQ971319 EFA00462.1 KQ461190 KPJ07188.1 NNAY01001418 OXU24046.1 AK417854 BAN21069.1 KK854035 PTY09925.1 KZ288310 PBC28605.1 GBYB01011216 JAG80983.1 GFDF01010908 JAV03176.1 AJVK01033360 GFXV01007941 MBW19746.1 GGMS01009354 MBY78557.1 ACPB03008668 GAHY01000807 JAA76703.1 ABLF02032059 GFTR01002792 JAW13634.1 KQ435119 KZC14719.1 GFDL01006410 JAV28635.1 AXCN02001078 DS231901 EDS44879.1 CH477272 EAT45227.1 ATLV01023814 KE525347 KFB49661.1 APGK01057455 BT128089 KB741280 KB632033 AEE63050.1 ENN70916.1 ERL88209.1 JRES01000502 KNC30601.1 APCN01003277 AAAB01008986 EAA00119.1 CH940647 EDW70816.1 CH933809 EDW19203.1 CH916366 EDV97790.1 JXUM01015999 JXUM01042477 GAPW01005163 KQ561326 KQ560445 JAC08435.1 KXJ78977.1 KXJ82384.1 AXCM01006131 CCAG010009396 JXJN01004648 GDKW01001771 JAI54824.1 GGFK01012031 MBW45352.1 GGFJ01010031 MBW59172.1 ADMH02001151 ETN63891.1 GDAI01002904 JAI14699.1 GL437917 EFN69899.1 CH963847 EDW73331.1 OUUW01000002 SPP77276.1 CH379069 EAL29746.1 CH479199 EDW29601.1 GAMC01006355 JAC00201.1 CH902618 EDV41074.1 CP012525 ALC44650.1 GL768221 EFZ11721.1 CH480844 EDW49854.1 CH954178 EDV52438.1 CM000363 CM002912 EDX11152.1 KMZ00663.1 AE014296 BT099881 AAF49088.1 ACX30043.1 AGB94775.1 CM000159 EDW95250.1 EDW95251.1 CAQQ02167088 KK107109 EZA59051.1 KQ981864 LKEZ01003687 KYN34190.1 KYN50698.1 KQ979568 KYN20959.1 GL888818 EGI57965.1 KQ976580 KYM79901.1 KQ982335 KYQ57608.1 UFQT01001854 SSX32019.1 ADTU01014890 GDIQ01210018 GDIP01076117 LRGB01000024 JAK41707.1 JAM27598.1 KZS21599.1 GL732525 EFX88651.1 QOIP01000008 RLU19937.1 KQ977985 KYM98350.1 KK114439 KFM62432.1 GDIP01015223 JAM88492.1 GANO01000818 JAB59053.1 JH431789
SOQ58915.1 NWSH01000601 PCG75408.1 GEZM01000267 JAV98444.1 GEBQ01020180 JAT19797.1 PYGN01001825 PSN32649.1 GEDC01000279 JAS37019.1 KK853239 KDR09495.1 KQ414583 KOC70743.1 KQ971319 EFA00462.1 KQ461190 KPJ07188.1 NNAY01001418 OXU24046.1 AK417854 BAN21069.1 KK854035 PTY09925.1 KZ288310 PBC28605.1 GBYB01011216 JAG80983.1 GFDF01010908 JAV03176.1 AJVK01033360 GFXV01007941 MBW19746.1 GGMS01009354 MBY78557.1 ACPB03008668 GAHY01000807 JAA76703.1 ABLF02032059 GFTR01002792 JAW13634.1 KQ435119 KZC14719.1 GFDL01006410 JAV28635.1 AXCN02001078 DS231901 EDS44879.1 CH477272 EAT45227.1 ATLV01023814 KE525347 KFB49661.1 APGK01057455 BT128089 KB741280 KB632033 AEE63050.1 ENN70916.1 ERL88209.1 JRES01000502 KNC30601.1 APCN01003277 AAAB01008986 EAA00119.1 CH940647 EDW70816.1 CH933809 EDW19203.1 CH916366 EDV97790.1 JXUM01015999 JXUM01042477 GAPW01005163 KQ561326 KQ560445 JAC08435.1 KXJ78977.1 KXJ82384.1 AXCM01006131 CCAG010009396 JXJN01004648 GDKW01001771 JAI54824.1 GGFK01012031 MBW45352.1 GGFJ01010031 MBW59172.1 ADMH02001151 ETN63891.1 GDAI01002904 JAI14699.1 GL437917 EFN69899.1 CH963847 EDW73331.1 OUUW01000002 SPP77276.1 CH379069 EAL29746.1 CH479199 EDW29601.1 GAMC01006355 JAC00201.1 CH902618 EDV41074.1 CP012525 ALC44650.1 GL768221 EFZ11721.1 CH480844 EDW49854.1 CH954178 EDV52438.1 CM000363 CM002912 EDX11152.1 KMZ00663.1 AE014296 BT099881 AAF49088.1 ACX30043.1 AGB94775.1 CM000159 EDW95250.1 EDW95251.1 CAQQ02167088 KK107109 EZA59051.1 KQ981864 LKEZ01003687 KYN34190.1 KYN50698.1 KQ979568 KYN20959.1 GL888818 EGI57965.1 KQ976580 KYM79901.1 KQ982335 KYQ57608.1 UFQT01001854 SSX32019.1 ADTU01014890 GDIQ01210018 GDIP01076117 LRGB01000024 JAK41707.1 JAM27598.1 KZS21599.1 GL732525 EFX88651.1 QOIP01000008 RLU19937.1 KQ977985 KYM98350.1 KK114439 KFM62432.1 GDIP01015223 JAM88492.1 GANO01000818 JAB59053.1 JH431789
Proteomes
UP000005204
UP000053268
UP000218220
UP000245037
UP000192223
UP000027135
+ More
UP000053825 UP000005203 UP000007266 UP000053240 UP000215335 UP000002358 UP000242457 UP000092462 UP000015103 UP000007819 UP000079169 UP000076502 UP000075886 UP000002320 UP000008820 UP000030765 UP000019118 UP000030742 UP000095301 UP000037069 UP000076407 UP000076408 UP000075840 UP000075901 UP000075903 UP000075902 UP000075882 UP000007062 UP000008792 UP000009192 UP000095300 UP000001070 UP000075881 UP000069940 UP000249989 UP000075883 UP000092444 UP000092443 UP000078200 UP000092460 UP000075920 UP000092445 UP000091820 UP000075885 UP000000673 UP000000311 UP000007798 UP000268350 UP000001819 UP000008744 UP000007801 UP000092553 UP000192221 UP000001292 UP000008711 UP000000304 UP000000803 UP000002282 UP000015102 UP000053097 UP000078541 UP000078492 UP000007755 UP000078540 UP000075809 UP000075900 UP000005205 UP000076858 UP000000305 UP000279307 UP000078542 UP000054359 UP000075884
UP000053825 UP000005203 UP000007266 UP000053240 UP000215335 UP000002358 UP000242457 UP000092462 UP000015103 UP000007819 UP000079169 UP000076502 UP000075886 UP000002320 UP000008820 UP000030765 UP000019118 UP000030742 UP000095301 UP000037069 UP000076407 UP000076408 UP000075840 UP000075901 UP000075903 UP000075902 UP000075882 UP000007062 UP000008792 UP000009192 UP000095300 UP000001070 UP000075881 UP000069940 UP000249989 UP000075883 UP000092444 UP000092443 UP000078200 UP000092460 UP000075920 UP000092445 UP000091820 UP000075885 UP000000673 UP000000311 UP000007798 UP000268350 UP000001819 UP000008744 UP000007801 UP000092553 UP000192221 UP000001292 UP000008711 UP000000304 UP000000803 UP000002282 UP000015102 UP000053097 UP000078541 UP000078492 UP000007755 UP000078540 UP000075809 UP000075900 UP000005205 UP000076858 UP000000305 UP000279307 UP000078542 UP000054359 UP000075884
PRIDE
Pfam
Interpro
SUPFAM
SSF53448
SSF53448
Gene 3D
ProteinModelPortal
H9JQ80
S4NW74
A0A194PVD4
A0A2H1X0P3
A0A2A4JUN5
A0A1Y1NKJ7
+ More
A0A1B6L7W2 A0A2P8XKX0 A0A1B6EGH6 A0A1W4X8Z3 A0A067QMS5 A0A0L7RIV7 A0A088AHU8 D6WF05 A0A194QNT6 A0A232EZV7 K7IYN0 R4WDU3 A0A2R7VVG0 A0A2A3EA52 A0A0C9RUD9 A0A1L8D9N0 A0A1B0EZ85 A0A2H8TZP6 A0A2S2QLD0 R4G8K6 J9K0V3 A0A224XRA5 A0A1S3DK63 A0A154PS35 A0A1Q3FM68 A0A182QK32 B0WDU2 Q17FB9 A0A084WHG7 J3JXW7 A0A1I8M150 A0A0L0CEC6 A0A2C9H8P1 A0A182Y207 A0A2C9GQU6 A0A182SA86 A0A182UM93 A0A182U4L7 A0A182KSX6 Q7Q020 B4LBK6 B4L0V3 A0A1I8PBL6 B4IZL5 A0A182JXQ0 A0A023EHT8 A0A182MCP6 A0A1B0FE16 A0A1A9Y2S1 A0A1A9VKV4 A0A1B0AWC7 A0A182WR14 A0A1B0A687 A0A1A9WSS8 A0A0N7Z920 A0A182P1R3 A0A2M4AXC0 A0A2M4C1I8 W5JIF5 A0A0K8TKB7 E2A9K9 B4MMI6 A0A3B0JV77 Q2LYW3 B4H024 W8BFY0 B3M8S3 A0A0M4F0H8 A0A1W4W6U6 E9J5Y1 B4IIT3 B3NE14 B4QRD9 Q9VW61 B4PFV3 T1GL01 A0A026WTR4 A0A151K4E5 A0A195E808 F4X6K5 A0A151I176 A0A151XBD5 A0A336MSD7 A0A182RQV3 A0A158NGF7 A0A0P5KGR4 E9FW33 A0A3L8DHP5 A0A151ID99 A0A087TBE3 A0A0N8DNX8 U5EVX3 A0A182N2M3 T1J1T7
A0A1B6L7W2 A0A2P8XKX0 A0A1B6EGH6 A0A1W4X8Z3 A0A067QMS5 A0A0L7RIV7 A0A088AHU8 D6WF05 A0A194QNT6 A0A232EZV7 K7IYN0 R4WDU3 A0A2R7VVG0 A0A2A3EA52 A0A0C9RUD9 A0A1L8D9N0 A0A1B0EZ85 A0A2H8TZP6 A0A2S2QLD0 R4G8K6 J9K0V3 A0A224XRA5 A0A1S3DK63 A0A154PS35 A0A1Q3FM68 A0A182QK32 B0WDU2 Q17FB9 A0A084WHG7 J3JXW7 A0A1I8M150 A0A0L0CEC6 A0A2C9H8P1 A0A182Y207 A0A2C9GQU6 A0A182SA86 A0A182UM93 A0A182U4L7 A0A182KSX6 Q7Q020 B4LBK6 B4L0V3 A0A1I8PBL6 B4IZL5 A0A182JXQ0 A0A023EHT8 A0A182MCP6 A0A1B0FE16 A0A1A9Y2S1 A0A1A9VKV4 A0A1B0AWC7 A0A182WR14 A0A1B0A687 A0A1A9WSS8 A0A0N7Z920 A0A182P1R3 A0A2M4AXC0 A0A2M4C1I8 W5JIF5 A0A0K8TKB7 E2A9K9 B4MMI6 A0A3B0JV77 Q2LYW3 B4H024 W8BFY0 B3M8S3 A0A0M4F0H8 A0A1W4W6U6 E9J5Y1 B4IIT3 B3NE14 B4QRD9 Q9VW61 B4PFV3 T1GL01 A0A026WTR4 A0A151K4E5 A0A195E808 F4X6K5 A0A151I176 A0A151XBD5 A0A336MSD7 A0A182RQV3 A0A158NGF7 A0A0P5KGR4 E9FW33 A0A3L8DHP5 A0A151ID99 A0A087TBE3 A0A0N8DNX8 U5EVX3 A0A182N2M3 T1J1T7
Ontologies
PANTHER
Topology
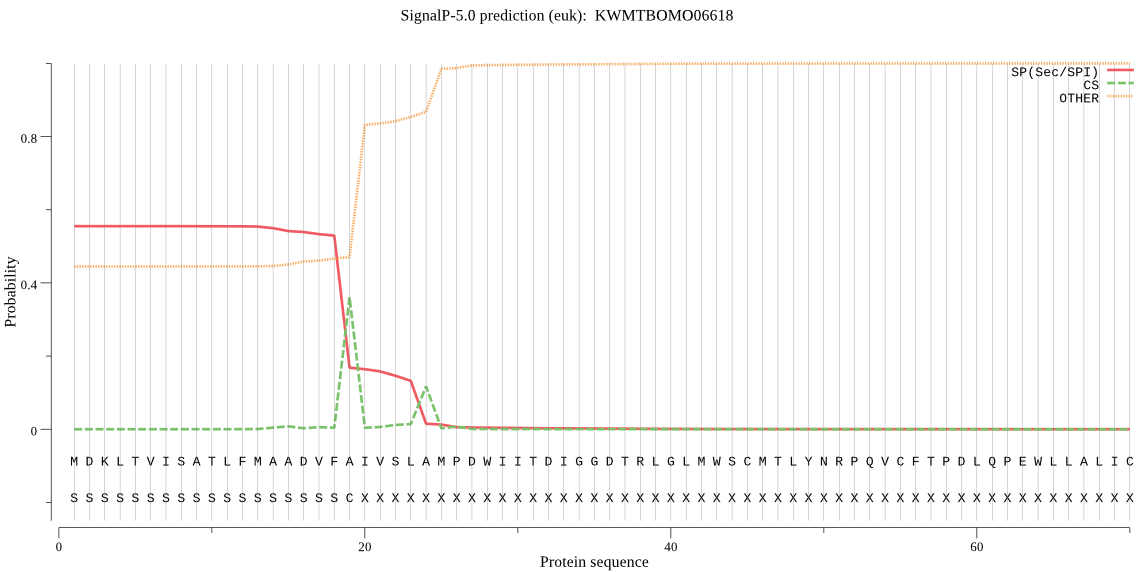
SignalP
Position: 1 - 19,
Likelihood: 0.554832
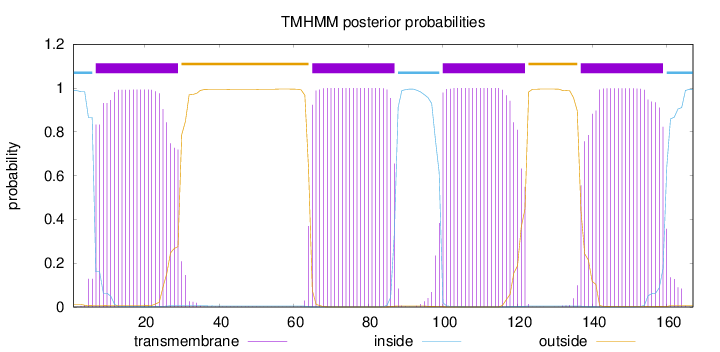
Length:
167
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
89.85493
Exp number, first 60 AAs:
22.01441
Total prob of N-in:
0.98831
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 64
TMhelix
65 - 87
inside
88 - 99
TMhelix
100 - 122
outside
123 - 136
TMhelix
137 - 159
inside
160 - 167
Population Genetic Test Statistics
Pi
144.764848
Theta
151.213313
Tajima's D
-0.044899
CLR
164.050827
CSRT
0.357082145892705
Interpretation
Uncertain
