Gene
KWMTBOMO06614
Pre Gene Modal
BGIBMGA011683
Annotation
Sodium/potassium-transporting_ATPase_subunit_alpha_[Papilio_xuthus]
Full name
Sodium/potassium-transporting ATPase subunit alpha
Alternative Name
Sodium pump subunit alpha
Location in the cell
Extracellular Reliability : 1.936
Sequence
CDS
ATGCAGACAATAATTTTTGTGATCGGTATCATTGTTGCGAATATTCCTGAAGGCTTACAACCTACAGTAACAGCTTCGCTAACGTTAACGGCTCAGCTAATGGTGAAGAAGCAATGTCTTGTTAAGAACTTAGATGCGATAGAGGCTTTAGGAGCATGCACTACCATTTGCTCGGATAAAACTGGTACTTTAACTCAGAACAAGATGTGTGTACATCACATTTGGATGTGGAACCAAACATACAATGTGTCAAAGATAGTTTTCCAGAGTTTGACGATATGTGGAGCTCTCTGTAGTACCGCTAGCATTCAAAGTGATGGTAATATAAGCGGCGATGCCTCAGAAAAAGCTATTTTAAATTTTTTGATGGAGCACGACAATCCAATGACCATTCGTGATAATTTTCCTAAAGTCGCAGAAATACCTTTTAATTCTGCAAATAAGTACCAAGCCAGCGTATGTCTTAACAAATATGATTCAAAATTTGTAGTACTAATGAAAGGAGCACCTGAATATATAGCCAATCGATGTGCTACCATCGCCCTAAAAACAGGAAATACCTCATTAACAACGGAAATGATGAAAGTGGTAGAATTAGCTACTGAAAAACTGGCCAACAGCGGTGAGAGGGTGTTAGCATTTGCTGATTTAACATTGAATACGAAAGAATTTCCAATTGACTTTGAATTTGACATAGATAATATTAATTTTCCACTGGACAAACTACGATTTTTAGGACTTTTAGGACTTATGGATCCTCCTAGACAAGAGGCAAGTTTGAAGTAG
Protein
MQTIIFVIGIIVANIPEGLQPTVTASLTLTAQLMVKKQCLVKNLDAIEALGACTTICSDKTGTLTQNKMCVHHIWMWNQTYNVSKIVFQSLTICGALCSTASIQSDGNISGDASEKAILNFLMEHDNPMTIRDNFPKVAEIPFNSANKYQASVCLNKYDSKFVVLMKGAPEYIANRCATIALKTGNTSLTTEMMKVVELATEKLANSGERVLAFADLTLNTKEFPIDFEFDIDNINFPLDKLRFLGLLGLMDPPRQEASLK
Summary
Catalytic Activity
ATP + H2O + K(+)(out) + Na(+)(in) = ADP + H(+) + K(+)(in) + Na(+)(out) + phosphate
Subunit
The sodium/potassium-transporting ATPase is composed of a catalytic alpha subunit, an auxiliary non-catalytic beta subunit and an additional regulatory subunit.
Similarity
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IIC subfamily.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IIC subfamily.
Keywords
ATP-binding
Cell membrane
Direct protein sequencing
Ion transport
Magnesium
Membrane
Metal-binding
Nucleotide-binding
Phosphoprotein
Potassium
Potassium transport
Sodium
Sodium transport
Translocase
Transmembrane
Transmembrane helix
Transport
Feature
chain Sodium/potassium-transporting ATPase subunit alpha
Uniprot
A0A2H1X0Q4
A0A2A4JVG5
A0A194PVC9
A0A3S2L7T1
A0A194QNU1
A0A212FJH3
+ More
A0A0L7LLV5 A0A3B3C748 A0A3B3C542 A0A3B3DJ94 A0A0S7EVS2 A0A2A6D3E5 A0A0S7F4F7 A0A077D1P7 A0A146Z6G7 A0A3P9HTK2 A0A3Q0S5D9 A0A077D5K3 A0A3P8UIA1 A8VUU1 W5MZI4 A0A093IWW3 I6SBF7 A0A3P9JAD5 A0A146RXH2 A0A146RWM0 J9ZVP5 I4AZ64 P05025 C6YYS9 A0A1I7RVE5 A0A146Z633 A0A2H4TJD7 C6YYT2 E2DV11 A0A091PGQ2 A0A1T4J336 A0A0P7X449 A0A077D328 I4AZ66 I4AZ95 A0A1T4J318 A0A0B4PKQ8 A0A0B4PLG0 C6YYS4 A0A3Q1IRH7 A0A093GA23 I6QED3 I4AZC7 C6YYS5 A0A077D2Y0 A0A0B4PL97 C6YYT3 A0A0A7A7P1 A0A2H2IJ96 A0A091FW86 A0A1T4J322 A8VUY5 A0A1T4J328 C6YYT5 B6C823 B6C821 E2DV18 A0A2H4TJG0 I4AZ59 B6C833 C6YYT4 B6C806 A0A3Q2DJP5 A0A3P8SRR3 A0A077D4W0 A0A077CZZ0 A0A3Q1G984 A0A093EG36 A0A164WKM6 F6LGU8 A0A159UTH1 A0A164WJT9 I4AZB4 A0A3Q1CJ40 A0A172R0I6 A0A3Q2DI97 A0A3Q4MRU1 K9MFB1 I4AZ81 A0A172R0I3 A0A146Q8B1 L7P4N9 A0A172R0I9 L7P4M9 A0A1B6FCR8 A0A172R0J9 A0A3S2MGA5 L7P5I7 C6YYU4 C6YYS6 A0A172R0J0 B6C817 B6C810 B6C813
A0A0L7LLV5 A0A3B3C748 A0A3B3C542 A0A3B3DJ94 A0A0S7EVS2 A0A2A6D3E5 A0A0S7F4F7 A0A077D1P7 A0A146Z6G7 A0A3P9HTK2 A0A3Q0S5D9 A0A077D5K3 A0A3P8UIA1 A8VUU1 W5MZI4 A0A093IWW3 I6SBF7 A0A3P9JAD5 A0A146RXH2 A0A146RWM0 J9ZVP5 I4AZ64 P05025 C6YYS9 A0A1I7RVE5 A0A146Z633 A0A2H4TJD7 C6YYT2 E2DV11 A0A091PGQ2 A0A1T4J336 A0A0P7X449 A0A077D328 I4AZ66 I4AZ95 A0A1T4J318 A0A0B4PKQ8 A0A0B4PLG0 C6YYS4 A0A3Q1IRH7 A0A093GA23 I6QED3 I4AZC7 C6YYS5 A0A077D2Y0 A0A0B4PL97 C6YYT3 A0A0A7A7P1 A0A2H2IJ96 A0A091FW86 A0A1T4J322 A8VUY5 A0A1T4J328 C6YYT5 B6C823 B6C821 E2DV18 A0A2H4TJG0 I4AZ59 B6C833 C6YYT4 B6C806 A0A3Q2DJP5 A0A3P8SRR3 A0A077D4W0 A0A077CZZ0 A0A3Q1G984 A0A093EG36 A0A164WKM6 F6LGU8 A0A159UTH1 A0A164WJT9 I4AZB4 A0A3Q1CJ40 A0A172R0I6 A0A3Q2DI97 A0A3Q4MRU1 K9MFB1 I4AZ81 A0A172R0I3 A0A146Q8B1 L7P4N9 A0A172R0I9 L7P4M9 A0A1B6FCR8 A0A172R0J9 A0A3S2MGA5 L7P5I7 C6YYU4 C6YYS6 A0A172R0J0 B6C817 B6C810 B6C813
EC Number
7.2.2.13
Pubmed
EMBL
ODYU01012538
SOQ58909.1
NWSH01000601
PCG75400.1
KQ459590
KPI97351.1
+ More
RSAL01000097 RVE47665.1 KQ461190 KPJ07193.1 AGBW02008266 OWR53886.1 JTDY01000701 KOB76166.1 GBYX01472542 GBYX01472530 GBYX01472503 GBYX01472490 JAO09151.1 ABKE03000003 PDM84919.1 GBYX01472539 GBYX01472522 GBYX01472493 GBYX01472483 JAO09161.1 KF935928 AIL26358.1 GCES01024474 JAR61849.1 KF935847 AIL26277.1 EU184744 ABW38135.1 AHAT01034984 AHAT01034985 KK564131 KFW03259.1 JQ885970 AFM73920.1 GCES01113576 JAQ72746.1 GCES01113577 JAQ72745.1 JX508624 AB759891 AFS60173.1 BAN17690.1 JX090933 AFM10183.1 X02810 EF646435 ABV59042.1 GCES01024473 JAR61850.1 MG250924 ATY93814.1 EF646438 ABV59045.1 GU245203 ADN93769.1 KK658884 KFQ06840.1 LT795110 SJX66706.1 JARO02002942 KPP71523.1 KF935915 AIL26345.1 JX090935 JX090973 AFM10185.1 AFM10223.1 JX090934 JX090964 AFM10184.1 AFM10214.1 LT795077 SJX66683.1 KJ434888 AIS74971.1 KJ434807 AIS74890.1 EF646430 ABV59037.1 KL215658 KFV66013.1 JN180941 AFK29493.1 JX090996 AFM10246.1 EF646431 ABV59038.1 KF935840 AIL26270.1 KJ434881 AIS74964.1 EF646439 ABV59046.1 KF649219 AHB86586.1 KL447417 KFO73311.1 LT795078 SJX66684.1 EU184754 ABW38145.1 LT795083 SJX66689.1 EF646441 ABV59048.1 EF646405 ABV57440.1 EF646403 ABV57438.1 GU245210 ADN93776.1 MG250941 ATY93831.1 JX090928 AFM10178.1 EF646415 ABV57450.1 EF646440 ABV59047.1 EF646388 ABV57423.1 KF935916 AIL26346.1 KF935839 KF935907 AIL26269.1 KL243832 KFV13486.1 KU976212 AMS36800.1 HQ995989 AEE80911.1 KU976197 AMS36785.1 KU976182 AMS36770.1 JX090983 AFM10233.1 KT318960 ANE06785.1 JQ279339 AFV15610.1 JX090950 AFM10200.1 KT318963 ANE06788.1 GCES01134068 JAQ52254.1 JN882407 JN882408 JN882409 JN882410 JN882411 JN882412 JN882413 JN882414 JN882415 JN882416 JN882417 JN882418 JN882419 JN882420 JN882421 JN882422 JN882423 JN882424 JN882425 JN882426 JN882427 JN882428 JN882429 JN882430 JN882431 JN882434 JN882435 JN882436 JN882437 JN882438 JN882439 JN882440 KT318961 AFK91637.1 AFK91638.1 AFK91639.1 AFK91640.1 AFK91641.1 AFK91642.1 AFK91643.1 AFK91644.1 AFK91645.1 AFK91646.1 AFK91647.1 AFK91648.1 AFK91649.1 AFK91650.1 AFK91651.1 AFK91652.1 AFK91653.1 AFK91654.1 AFK91655.1 AFK91656.1 AFK91657.1 AFK91658.1 AFK91659.1 AFK91660.1 AFK91661.1 AFK91664.1 AFK91665.1 AFK91666.1 AFK91667.1 AFK91668.1 AFK91669.1 AFK91670.1 ANE06786.1 KT318959 KT318965 KT318966 ANE06784.1 ANE06790.1 ANE06791.1 JN882406 JN882432 JN882433 KT318950 KT318951 KT318952 KT318953 KT318954 KT318955 KT318956 KT318957 KT318958 AFK91636.1 AFK91662.1 AFK91663.1 ANE06775.1 ANE06776.1 ANE06777.1 ANE06778.1 ANE06779.1 ANE06780.1 ANE06781.1 ANE06782.1 ANE06783.1 GECZ01021753 JAS48016.1 KT318967 ANE06792.1 CM012438 RVE75278.1 JN882377 AFK91635.1 EF646450 ABV59057.1 EF646432 ABV59039.1 KT318962 ANE06787.1 EF646399 ABV57434.1 EF646392 ABV57427.1 EF646395 ABV57430.1
RSAL01000097 RVE47665.1 KQ461190 KPJ07193.1 AGBW02008266 OWR53886.1 JTDY01000701 KOB76166.1 GBYX01472542 GBYX01472530 GBYX01472503 GBYX01472490 JAO09151.1 ABKE03000003 PDM84919.1 GBYX01472539 GBYX01472522 GBYX01472493 GBYX01472483 JAO09161.1 KF935928 AIL26358.1 GCES01024474 JAR61849.1 KF935847 AIL26277.1 EU184744 ABW38135.1 AHAT01034984 AHAT01034985 KK564131 KFW03259.1 JQ885970 AFM73920.1 GCES01113576 JAQ72746.1 GCES01113577 JAQ72745.1 JX508624 AB759891 AFS60173.1 BAN17690.1 JX090933 AFM10183.1 X02810 EF646435 ABV59042.1 GCES01024473 JAR61850.1 MG250924 ATY93814.1 EF646438 ABV59045.1 GU245203 ADN93769.1 KK658884 KFQ06840.1 LT795110 SJX66706.1 JARO02002942 KPP71523.1 KF935915 AIL26345.1 JX090935 JX090973 AFM10185.1 AFM10223.1 JX090934 JX090964 AFM10184.1 AFM10214.1 LT795077 SJX66683.1 KJ434888 AIS74971.1 KJ434807 AIS74890.1 EF646430 ABV59037.1 KL215658 KFV66013.1 JN180941 AFK29493.1 JX090996 AFM10246.1 EF646431 ABV59038.1 KF935840 AIL26270.1 KJ434881 AIS74964.1 EF646439 ABV59046.1 KF649219 AHB86586.1 KL447417 KFO73311.1 LT795078 SJX66684.1 EU184754 ABW38145.1 LT795083 SJX66689.1 EF646441 ABV59048.1 EF646405 ABV57440.1 EF646403 ABV57438.1 GU245210 ADN93776.1 MG250941 ATY93831.1 JX090928 AFM10178.1 EF646415 ABV57450.1 EF646440 ABV59047.1 EF646388 ABV57423.1 KF935916 AIL26346.1 KF935839 KF935907 AIL26269.1 KL243832 KFV13486.1 KU976212 AMS36800.1 HQ995989 AEE80911.1 KU976197 AMS36785.1 KU976182 AMS36770.1 JX090983 AFM10233.1 KT318960 ANE06785.1 JQ279339 AFV15610.1 JX090950 AFM10200.1 KT318963 ANE06788.1 GCES01134068 JAQ52254.1 JN882407 JN882408 JN882409 JN882410 JN882411 JN882412 JN882413 JN882414 JN882415 JN882416 JN882417 JN882418 JN882419 JN882420 JN882421 JN882422 JN882423 JN882424 JN882425 JN882426 JN882427 JN882428 JN882429 JN882430 JN882431 JN882434 JN882435 JN882436 JN882437 JN882438 JN882439 JN882440 KT318961 AFK91637.1 AFK91638.1 AFK91639.1 AFK91640.1 AFK91641.1 AFK91642.1 AFK91643.1 AFK91644.1 AFK91645.1 AFK91646.1 AFK91647.1 AFK91648.1 AFK91649.1 AFK91650.1 AFK91651.1 AFK91652.1 AFK91653.1 AFK91654.1 AFK91655.1 AFK91656.1 AFK91657.1 AFK91658.1 AFK91659.1 AFK91660.1 AFK91661.1 AFK91664.1 AFK91665.1 AFK91666.1 AFK91667.1 AFK91668.1 AFK91669.1 AFK91670.1 ANE06786.1 KT318959 KT318965 KT318966 ANE06784.1 ANE06790.1 ANE06791.1 JN882406 JN882432 JN882433 KT318950 KT318951 KT318952 KT318953 KT318954 KT318955 KT318956 KT318957 KT318958 AFK91636.1 AFK91662.1 AFK91663.1 ANE06775.1 ANE06776.1 ANE06777.1 ANE06778.1 ANE06779.1 ANE06780.1 ANE06781.1 ANE06782.1 ANE06783.1 GECZ01021753 JAS48016.1 KT318967 ANE06792.1 CM012438 RVE75278.1 JN882377 AFK91635.1 EF646450 ABV59057.1 EF646432 ABV59039.1 KT318962 ANE06787.1 EF646399 ABV57434.1 EF646392 ABV57427.1 EF646395 ABV57430.1
Proteomes
PRIDE
Interpro
IPR023214
HAD_sf
+ More
IPR036412 HAD-like_sf
IPR023299 ATPase_P-typ_cyto_dom_N
IPR023298 ATPase_P-typ_TM_dom_sf
IPR006068 ATPase_P-typ_cation-transptr_C
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR001757 P_typ_ATPase
IPR004014 ATPase_P-typ_cation-transptr_N
IPR018303 ATPase_P-typ_P_site
IPR005775 P-type_ATPase_IIC
IPR036412 HAD-like_sf
IPR023299 ATPase_P-typ_cyto_dom_N
IPR023298 ATPase_P-typ_TM_dom_sf
IPR006068 ATPase_P-typ_cation-transptr_C
IPR008250 ATPase_P-typ_transduc_dom_A_sf
IPR001757 P_typ_ATPase
IPR004014 ATPase_P-typ_cation-transptr_N
IPR018303 ATPase_P-typ_P_site
IPR005775 P-type_ATPase_IIC
Gene 3D
ProteinModelPortal
A0A2H1X0Q4
A0A2A4JVG5
A0A194PVC9
A0A3S2L7T1
A0A194QNU1
A0A212FJH3
+ More
A0A0L7LLV5 A0A3B3C748 A0A3B3C542 A0A3B3DJ94 A0A0S7EVS2 A0A2A6D3E5 A0A0S7F4F7 A0A077D1P7 A0A146Z6G7 A0A3P9HTK2 A0A3Q0S5D9 A0A077D5K3 A0A3P8UIA1 A8VUU1 W5MZI4 A0A093IWW3 I6SBF7 A0A3P9JAD5 A0A146RXH2 A0A146RWM0 J9ZVP5 I4AZ64 P05025 C6YYS9 A0A1I7RVE5 A0A146Z633 A0A2H4TJD7 C6YYT2 E2DV11 A0A091PGQ2 A0A1T4J336 A0A0P7X449 A0A077D328 I4AZ66 I4AZ95 A0A1T4J318 A0A0B4PKQ8 A0A0B4PLG0 C6YYS4 A0A3Q1IRH7 A0A093GA23 I6QED3 I4AZC7 C6YYS5 A0A077D2Y0 A0A0B4PL97 C6YYT3 A0A0A7A7P1 A0A2H2IJ96 A0A091FW86 A0A1T4J322 A8VUY5 A0A1T4J328 C6YYT5 B6C823 B6C821 E2DV18 A0A2H4TJG0 I4AZ59 B6C833 C6YYT4 B6C806 A0A3Q2DJP5 A0A3P8SRR3 A0A077D4W0 A0A077CZZ0 A0A3Q1G984 A0A093EG36 A0A164WKM6 F6LGU8 A0A159UTH1 A0A164WJT9 I4AZB4 A0A3Q1CJ40 A0A172R0I6 A0A3Q2DI97 A0A3Q4MRU1 K9MFB1 I4AZ81 A0A172R0I3 A0A146Q8B1 L7P4N9 A0A172R0I9 L7P4M9 A0A1B6FCR8 A0A172R0J9 A0A3S2MGA5 L7P5I7 C6YYU4 C6YYS6 A0A172R0J0 B6C817 B6C810 B6C813
A0A0L7LLV5 A0A3B3C748 A0A3B3C542 A0A3B3DJ94 A0A0S7EVS2 A0A2A6D3E5 A0A0S7F4F7 A0A077D1P7 A0A146Z6G7 A0A3P9HTK2 A0A3Q0S5D9 A0A077D5K3 A0A3P8UIA1 A8VUU1 W5MZI4 A0A093IWW3 I6SBF7 A0A3P9JAD5 A0A146RXH2 A0A146RWM0 J9ZVP5 I4AZ64 P05025 C6YYS9 A0A1I7RVE5 A0A146Z633 A0A2H4TJD7 C6YYT2 E2DV11 A0A091PGQ2 A0A1T4J336 A0A0P7X449 A0A077D328 I4AZ66 I4AZ95 A0A1T4J318 A0A0B4PKQ8 A0A0B4PLG0 C6YYS4 A0A3Q1IRH7 A0A093GA23 I6QED3 I4AZC7 C6YYS5 A0A077D2Y0 A0A0B4PL97 C6YYT3 A0A0A7A7P1 A0A2H2IJ96 A0A091FW86 A0A1T4J322 A8VUY5 A0A1T4J328 C6YYT5 B6C823 B6C821 E2DV18 A0A2H4TJG0 I4AZ59 B6C833 C6YYT4 B6C806 A0A3Q2DJP5 A0A3P8SRR3 A0A077D4W0 A0A077CZZ0 A0A3Q1G984 A0A093EG36 A0A164WKM6 F6LGU8 A0A159UTH1 A0A164WJT9 I4AZB4 A0A3Q1CJ40 A0A172R0I6 A0A3Q2DI97 A0A3Q4MRU1 K9MFB1 I4AZ81 A0A172R0I3 A0A146Q8B1 L7P4N9 A0A172R0I9 L7P4M9 A0A1B6FCR8 A0A172R0J9 A0A3S2MGA5 L7P5I7 C6YYU4 C6YYS6 A0A172R0J0 B6C817 B6C810 B6C813
PDB
5Y0B
E-value=3.08548e-44,
Score=447
Ontologies
GO
Topology
Subcellular location
Membrane
Cell membrane
Cell membrane
SignalP
Position: 1 - 18,
Likelihood: 0.713415
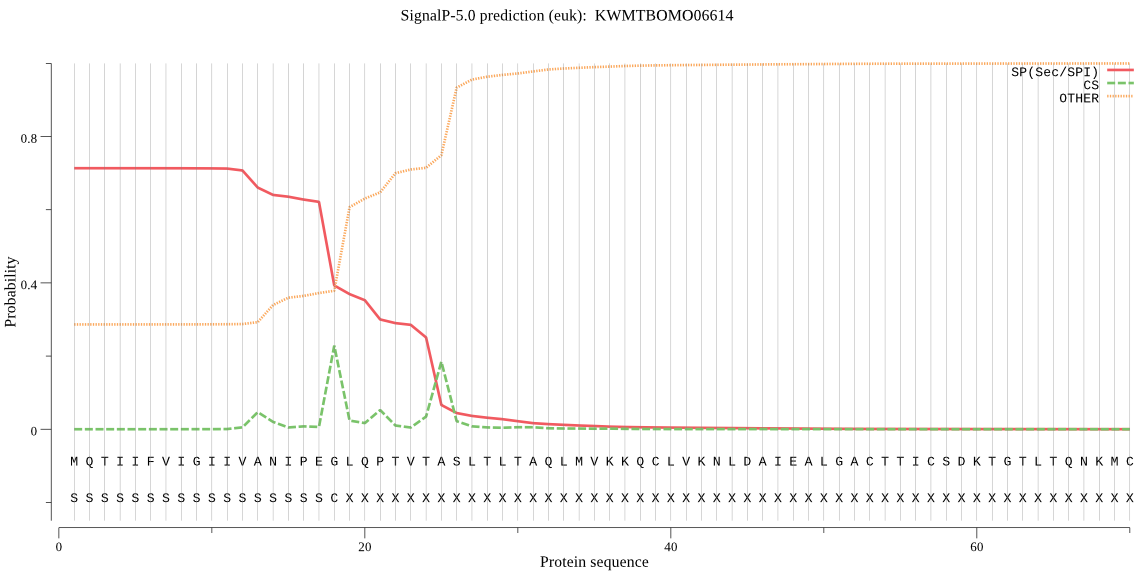
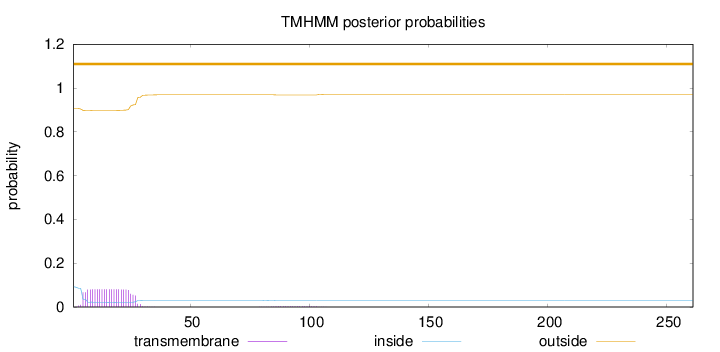
Length:
261
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.86347
Exp number, first 60 AAs:
1.80162
Total prob of N-in:
0.09271
outside
1 - 261
Population Genetic Test Statistics
Pi
146.416774
Theta
151.828449
Tajima's D
-0.112053
CLR
0.622056
CSRT
0.342382880855957
Interpretation
Uncertain
