Gene
KWMTBOMO06611 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011792
Annotation
PREDICTED:_TPPP_family_protein_CG45057-like_[Amyelois_transitella]
Full name
TPPP family protein CG45057
Location in the cell
Nuclear Reliability : 2.877
Sequence
CDS
ATGTCTACTGAAGCACAGAATACGGATGCCGCTGTGGAGCAAGTGACACAGGAAGTCAAGGACGTCAAGCTTGAGAACGGGAACGCGCCGGGAGCATCCAATGGAACGTCCAGTAAAAGCGAGGATAACGCCTTATCTTTCAAGGAAGCCTTCAAGGCGTTTTCCAAATTTGGAGATCCCAAGTCCGATGGAAAAGCCATCACGCTCTCGCAAAGCGACAAATGGATGAAGCAAGCCAAAGTCATTGATGGAAAGAAAATAACAACAACGGACACGGCCATTCACTTCAAAAAACTCAAATCGGTAAAACTCGGCATCGACGACTACCAGAAGTTTCTAGATGATCTCGCGAAGAACAAAAAAGTGGAACTTGACGAAATTAAGAAGAAGTTGACAACTTGCGGCCAACCGGGAATTACATCACACGTTACAAAATCACCGGCAGCCGCAGCGGCCGTAGATCGATTGACAGACACAAGCAAATATACTGGCTCGCACAGGCAGCGCTTCGATGAAACTGGAAAAGGGAAGGGAATCGCCGGCAGGAAAGACTTAGTCGACGGGTCCGGCTACGTCACTGGATACCAACATAAAGATACTTACAATAAATCTCACTAA
Protein
MSTEAQNTDAAVEQVTQEVKDVKLENGNAPGASNGTSSKSEDNALSFKEAFKAFSKFGDPKSDGKAITLSQSDKWMKQAKVIDGKKITTTDTAIHFKKLKSVKLGIDDYQKFLDDLAKNKKVELDEIKKKLTTCGQPGITSHVTKSPAAAAAVDRLTDTSKYTGSHRQRFDETGKGKGIAGRKDLVDGSGYVTGYQHKDTYNKSH
Summary
Similarity
Belongs to the TPPP family.
Keywords
Complete proteome
Reference proteome
Feature
chain TPPP family protein CG45057
Uniprot
H9JQI6
A0A1E1WQM0
A0A3S2NYA6
A0A194QUG6
A0A212FJJ3
A0A194PWG5
+ More
S4PTD1 E2A3C6 A0A1B0D5U9 D6WGQ8 A0A1L8DMB8 A0A1Y1L2A6 A0A0L7LS62 A0A1B6L712 B4HJL7 A0A1S4FJV3 M9NFZ9 Q9VV43 B4QME0 A0A1W4UWD6 B4PHI4 A0A0J9RWR6 A0A1Q3FPS1 B4ITP8 A0A0R1DZD1 Q16YE8 A0A0R1E818 A0A026WVW0 A0A1W4UH63 A0A3L8DGX8 B3NDS3 A0A182RJL9 A0A0R1DZE3 A0A0R1E7H2 A0A0Q5U5N8 A0A0R3P8V6 Q29EJ5 A0A084WEZ2 A0A0J7KKU9 B0WH59 A0A182I6R9 B4H238 A0A182MRS9 E2BTF0 A0A1J1I6Q4 A0A182QU40 A0A182L3L3 B4MX81 A0A3B0K337 A0A182N999 A0A182PII4 T1E2A3 A0A182WWI8 A0A0K8UVX4 A0A034WF27 B3M5K3 A0A182VE43 A0A182U6F0 A7URV3 W8B927 A0A034WDH6 Q7PT09 A0A0K8V8P4 A0A182FGT7 A0A2M4BZ15 A0A0P8XTR4 A0A0N0BK83 F4WRB2 A0A023EK52 A0A023EK42 A0A0A1XBK9 A0A0A1WRL3 W5JNY0 A0A2J7Q6I1 T1E2X3 A0A182JAI2 A0A069DWC4 V9ID76 A0A195BAL0 A0A158NUP8 N6TH68 A0A182H3P1 A0A182JP74 A0A1B6IV74 A0A1B0BY29 A0A151ISD4 A0A1B0A6E6 A0A0V0GAM1 B4J0U2 A0A1A9VIS6 A0A067QX45 A0A1W4W501 A0A2A3EJN7 A0A224XZY5 A0A023FCX5 D3TRI2 A0A0M4EE33 E2J7I6
S4PTD1 E2A3C6 A0A1B0D5U9 D6WGQ8 A0A1L8DMB8 A0A1Y1L2A6 A0A0L7LS62 A0A1B6L712 B4HJL7 A0A1S4FJV3 M9NFZ9 Q9VV43 B4QME0 A0A1W4UWD6 B4PHI4 A0A0J9RWR6 A0A1Q3FPS1 B4ITP8 A0A0R1DZD1 Q16YE8 A0A0R1E818 A0A026WVW0 A0A1W4UH63 A0A3L8DGX8 B3NDS3 A0A182RJL9 A0A0R1DZE3 A0A0R1E7H2 A0A0Q5U5N8 A0A0R3P8V6 Q29EJ5 A0A084WEZ2 A0A0J7KKU9 B0WH59 A0A182I6R9 B4H238 A0A182MRS9 E2BTF0 A0A1J1I6Q4 A0A182QU40 A0A182L3L3 B4MX81 A0A3B0K337 A0A182N999 A0A182PII4 T1E2A3 A0A182WWI8 A0A0K8UVX4 A0A034WF27 B3M5K3 A0A182VE43 A0A182U6F0 A7URV3 W8B927 A0A034WDH6 Q7PT09 A0A0K8V8P4 A0A182FGT7 A0A2M4BZ15 A0A0P8XTR4 A0A0N0BK83 F4WRB2 A0A023EK52 A0A023EK42 A0A0A1XBK9 A0A0A1WRL3 W5JNY0 A0A2J7Q6I1 T1E2X3 A0A182JAI2 A0A069DWC4 V9ID76 A0A195BAL0 A0A158NUP8 N6TH68 A0A182H3P1 A0A182JP74 A0A1B6IV74 A0A1B0BY29 A0A151ISD4 A0A1B0A6E6 A0A0V0GAM1 B4J0U2 A0A1A9VIS6 A0A067QX45 A0A1W4W501 A0A2A3EJN7 A0A224XZY5 A0A023FCX5 D3TRI2 A0A0M4EE33 E2J7I6
Pubmed
19121390
26354079
22118469
23622113
20798317
18362917
+ More
19820115 28004739 26227816 17994087 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 22936249 17550304 24508170 30249741 15632085 24438588 20966253 24330624 25348373 12364791 14747013 17210077 24495485 18057021 21719571 24945155 25830018 20920257 23761445 26334808 21347285 23537049 26483478 24845553 25474469 20353571
19820115 28004739 26227816 17994087 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 22936249 17550304 24508170 30249741 15632085 24438588 20966253 24330624 25348373 12364791 14747013 17210077 24495485 18057021 21719571 24945155 25830018 20920257 23761445 26334808 21347285 23537049 26483478 24845553 25474469 20353571
EMBL
BABH01012031
GDQN01001772
JAT89282.1
RSAL01000097
RVE47668.1
KQ461190
+ More
KPJ07196.1 AGBW02008266 OWR53879.1 KQ459590 KPI97348.1 GAIX01012248 JAA80312.1 GL436364 EFN72070.1 AJVK01000389 AJVK01000390 KQ971327 EEZ99609.1 GFDF01006471 JAV07613.1 GEZM01068530 JAV66908.1 JTDY01000254 KOB78041.1 GEBQ01020542 JAT19435.1 CH480815 EDW41744.1 AE014296 AFH04463.1 AGB94631.1 AGB94632.1 AY071493 AY071358 CM000363 CM002912 EDX10709.1 KMZ00027.1 CM000159 EDW95422.1 KMZ00028.1 GFDL01005444 JAV29601.1 CH891728 EDW99761.1 KRK02428.1 CH477517 EAT39656.1 KRK05225.1 KK107109 EZA59234.1 QOIP01000008 RLU19700.1 CH954178 EDV52277.1 KRK02427.1 KRK05224.1 KQS44206.1 CH379070 KRT08627.1 EAL30065.3 ATLV01023266 KE525341 KFB48786.1 LBMM01006125 KMQ90902.1 DS231933 EDS27566.1 APCN01003693 CH479203 EDW30390.1 AXCM01000934 GL450402 EFN81031.1 CVRI01000043 CRK95875.1 AXCN02000558 CH963876 EDW76914.1 OUUW01000012 SPP87072.1 GALA01001405 JAA93447.1 GDHF01021804 JAI30510.1 GAKP01006237 JAC52715.1 CH902618 EDV39613.1 AAAB01008807 EDO64745.1 GAMC01011408 JAB95147.1 GAKP01006238 JAC52714.1 EAA04714.5 GDHF01017010 GDHF01008668 JAI35304.1 JAI43646.1 GGFJ01009103 MBW58244.1 KPU78108.1 KPU78109.1 KQ435706 KOX80211.1 GL888284 EGI63329.1 GAPW01004101 JAC09497.1 GAPW01004100 JAC09498.1 GBXI01005962 JAD08330.1 GBXI01013137 JAD01155.1 ADMH02000528 ETN66092.1 NEVH01017474 PNF24186.1 GALA01000895 JAA93957.1 AXCP01008358 GBGD01003260 JAC85629.1 JR039218 JR039219 AEY58586.1 AEY58587.1 KQ976532 KYM81586.1 ADTU01026655 APGK01027109 KB740648 KB632292 ENN79779.1 ERL91604.1 JXUM01107734 KQ565169 KXJ71259.1 GECU01024608 GECU01016865 JAS83098.1 JAS90841.1 JXJN01022474 KQ981085 KYN09592.1 GECL01001248 JAP04876.1 CH916366 EDV95763.1 KK852855 KDR14913.1 KZ288235 PBC31396.1 GFTR01002765 JAW13661.1 GBBI01000014 JAC18698.1 EZ424034 ADD20310.1 CP012525 ALC43971.1 HP429404 ADN29904.1
KPJ07196.1 AGBW02008266 OWR53879.1 KQ459590 KPI97348.1 GAIX01012248 JAA80312.1 GL436364 EFN72070.1 AJVK01000389 AJVK01000390 KQ971327 EEZ99609.1 GFDF01006471 JAV07613.1 GEZM01068530 JAV66908.1 JTDY01000254 KOB78041.1 GEBQ01020542 JAT19435.1 CH480815 EDW41744.1 AE014296 AFH04463.1 AGB94631.1 AGB94632.1 AY071493 AY071358 CM000363 CM002912 EDX10709.1 KMZ00027.1 CM000159 EDW95422.1 KMZ00028.1 GFDL01005444 JAV29601.1 CH891728 EDW99761.1 KRK02428.1 CH477517 EAT39656.1 KRK05225.1 KK107109 EZA59234.1 QOIP01000008 RLU19700.1 CH954178 EDV52277.1 KRK02427.1 KRK05224.1 KQS44206.1 CH379070 KRT08627.1 EAL30065.3 ATLV01023266 KE525341 KFB48786.1 LBMM01006125 KMQ90902.1 DS231933 EDS27566.1 APCN01003693 CH479203 EDW30390.1 AXCM01000934 GL450402 EFN81031.1 CVRI01000043 CRK95875.1 AXCN02000558 CH963876 EDW76914.1 OUUW01000012 SPP87072.1 GALA01001405 JAA93447.1 GDHF01021804 JAI30510.1 GAKP01006237 JAC52715.1 CH902618 EDV39613.1 AAAB01008807 EDO64745.1 GAMC01011408 JAB95147.1 GAKP01006238 JAC52714.1 EAA04714.5 GDHF01017010 GDHF01008668 JAI35304.1 JAI43646.1 GGFJ01009103 MBW58244.1 KPU78108.1 KPU78109.1 KQ435706 KOX80211.1 GL888284 EGI63329.1 GAPW01004101 JAC09497.1 GAPW01004100 JAC09498.1 GBXI01005962 JAD08330.1 GBXI01013137 JAD01155.1 ADMH02000528 ETN66092.1 NEVH01017474 PNF24186.1 GALA01000895 JAA93957.1 AXCP01008358 GBGD01003260 JAC85629.1 JR039218 JR039219 AEY58586.1 AEY58587.1 KQ976532 KYM81586.1 ADTU01026655 APGK01027109 KB740648 KB632292 ENN79779.1 ERL91604.1 JXUM01107734 KQ565169 KXJ71259.1 GECU01024608 GECU01016865 JAS83098.1 JAS90841.1 JXJN01022474 KQ981085 KYN09592.1 GECL01001248 JAP04876.1 CH916366 EDV95763.1 KK852855 KDR14913.1 KZ288235 PBC31396.1 GFTR01002765 JAW13661.1 GBBI01000014 JAC18698.1 EZ424034 ADD20310.1 CP012525 ALC43971.1 HP429404 ADN29904.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000007151
UP000053268
UP000000311
+ More
UP000092462 UP000007266 UP000037510 UP000001292 UP000000803 UP000000304 UP000192221 UP000002282 UP000008820 UP000053097 UP000279307 UP000008711 UP000075900 UP000001819 UP000030765 UP000036403 UP000002320 UP000075840 UP000008744 UP000075883 UP000008237 UP000183832 UP000075886 UP000075882 UP000007798 UP000268350 UP000075884 UP000075885 UP000076407 UP000007801 UP000075903 UP000075902 UP000007062 UP000069272 UP000053105 UP000007755 UP000000673 UP000235965 UP000075880 UP000078540 UP000005205 UP000019118 UP000030742 UP000069940 UP000249989 UP000075881 UP000092460 UP000078492 UP000092445 UP000001070 UP000078200 UP000027135 UP000192223 UP000242457 UP000092553
UP000092462 UP000007266 UP000037510 UP000001292 UP000000803 UP000000304 UP000192221 UP000002282 UP000008820 UP000053097 UP000279307 UP000008711 UP000075900 UP000001819 UP000030765 UP000036403 UP000002320 UP000075840 UP000008744 UP000075883 UP000008237 UP000183832 UP000075886 UP000075882 UP000007798 UP000268350 UP000075884 UP000075885 UP000076407 UP000007801 UP000075903 UP000075902 UP000007062 UP000069272 UP000053105 UP000007755 UP000000673 UP000235965 UP000075880 UP000078540 UP000005205 UP000019118 UP000030742 UP000069940 UP000249989 UP000075881 UP000092460 UP000078492 UP000092445 UP000001070 UP000078200 UP000027135 UP000192223 UP000242457 UP000092553
Pfam
PF05517 p25-alpha
SUPFAM
SSF47473
SSF47473
ProteinModelPortal
H9JQI6
A0A1E1WQM0
A0A3S2NYA6
A0A194QUG6
A0A212FJJ3
A0A194PWG5
+ More
S4PTD1 E2A3C6 A0A1B0D5U9 D6WGQ8 A0A1L8DMB8 A0A1Y1L2A6 A0A0L7LS62 A0A1B6L712 B4HJL7 A0A1S4FJV3 M9NFZ9 Q9VV43 B4QME0 A0A1W4UWD6 B4PHI4 A0A0J9RWR6 A0A1Q3FPS1 B4ITP8 A0A0R1DZD1 Q16YE8 A0A0R1E818 A0A026WVW0 A0A1W4UH63 A0A3L8DGX8 B3NDS3 A0A182RJL9 A0A0R1DZE3 A0A0R1E7H2 A0A0Q5U5N8 A0A0R3P8V6 Q29EJ5 A0A084WEZ2 A0A0J7KKU9 B0WH59 A0A182I6R9 B4H238 A0A182MRS9 E2BTF0 A0A1J1I6Q4 A0A182QU40 A0A182L3L3 B4MX81 A0A3B0K337 A0A182N999 A0A182PII4 T1E2A3 A0A182WWI8 A0A0K8UVX4 A0A034WF27 B3M5K3 A0A182VE43 A0A182U6F0 A7URV3 W8B927 A0A034WDH6 Q7PT09 A0A0K8V8P4 A0A182FGT7 A0A2M4BZ15 A0A0P8XTR4 A0A0N0BK83 F4WRB2 A0A023EK52 A0A023EK42 A0A0A1XBK9 A0A0A1WRL3 W5JNY0 A0A2J7Q6I1 T1E2X3 A0A182JAI2 A0A069DWC4 V9ID76 A0A195BAL0 A0A158NUP8 N6TH68 A0A182H3P1 A0A182JP74 A0A1B6IV74 A0A1B0BY29 A0A151ISD4 A0A1B0A6E6 A0A0V0GAM1 B4J0U2 A0A1A9VIS6 A0A067QX45 A0A1W4W501 A0A2A3EJN7 A0A224XZY5 A0A023FCX5 D3TRI2 A0A0M4EE33 E2J7I6
S4PTD1 E2A3C6 A0A1B0D5U9 D6WGQ8 A0A1L8DMB8 A0A1Y1L2A6 A0A0L7LS62 A0A1B6L712 B4HJL7 A0A1S4FJV3 M9NFZ9 Q9VV43 B4QME0 A0A1W4UWD6 B4PHI4 A0A0J9RWR6 A0A1Q3FPS1 B4ITP8 A0A0R1DZD1 Q16YE8 A0A0R1E818 A0A026WVW0 A0A1W4UH63 A0A3L8DGX8 B3NDS3 A0A182RJL9 A0A0R1DZE3 A0A0R1E7H2 A0A0Q5U5N8 A0A0R3P8V6 Q29EJ5 A0A084WEZ2 A0A0J7KKU9 B0WH59 A0A182I6R9 B4H238 A0A182MRS9 E2BTF0 A0A1J1I6Q4 A0A182QU40 A0A182L3L3 B4MX81 A0A3B0K337 A0A182N999 A0A182PII4 T1E2A3 A0A182WWI8 A0A0K8UVX4 A0A034WF27 B3M5K3 A0A182VE43 A0A182U6F0 A7URV3 W8B927 A0A034WDH6 Q7PT09 A0A0K8V8P4 A0A182FGT7 A0A2M4BZ15 A0A0P8XTR4 A0A0N0BK83 F4WRB2 A0A023EK52 A0A023EK42 A0A0A1XBK9 A0A0A1WRL3 W5JNY0 A0A2J7Q6I1 T1E2X3 A0A182JAI2 A0A069DWC4 V9ID76 A0A195BAL0 A0A158NUP8 N6TH68 A0A182H3P1 A0A182JP74 A0A1B6IV74 A0A1B0BY29 A0A151ISD4 A0A1B0A6E6 A0A0V0GAM1 B4J0U2 A0A1A9VIS6 A0A067QX45 A0A1W4W501 A0A2A3EJN7 A0A224XZY5 A0A023FCX5 D3TRI2 A0A0M4EE33 E2J7I6
PDB
2JRF
E-value=2.00218e-19,
Score=232
Ontologies
PANTHER
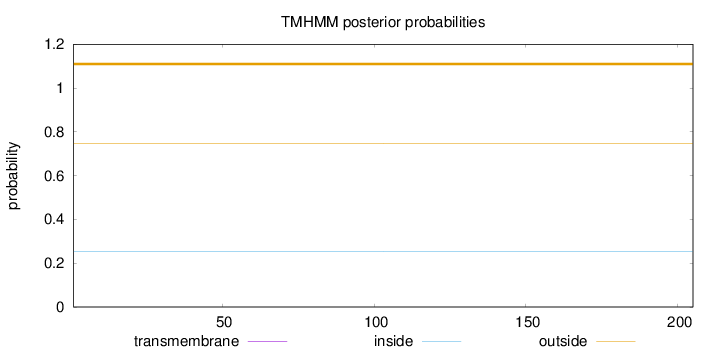
Topology
Length:
205
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00016
Exp number, first 60 AAs:
0
Total prob of N-in:
0.25215
outside
1 - 205
Population Genetic Test Statistics
Pi
253.377861
Theta
179.240412
Tajima's D
0.908555
CLR
0.570753
CSRT
0.630668466576671
Interpretation
Uncertain
