Gene
KWMTBOMO06608
Pre Gene Modal
BGIBMGA011794
Annotation
PREDICTED:_cirhin_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.213 PlasmaMembrane Reliability : 1.114
Sequence
CDS
ATGGCTACTAGCGTACATCGTATTCGTTATTATAATCCCCAACCAACCCCAGTGAATTGTATCGCTTATAATAAATTAAATAAACAATTGTCCCTCGCGAGGAAAGATTCAAGTATTGAAATATGGGACATGACACATTTCCCATTCCTCTTAAAATTCATACCTGCAGTAAAGGACGGCTCAGTAGAAGCTCTAGGCTGGGTGAAAGACAGACTGCTTACAACCGGATTAGGCGGTGCATTAGTTGAATGGAATCTTGAAGAAATGTGTATAAGAAATACAATATTATTAACTGGATATGCTGCTTGGTGTCTTGATGTTAGCCCAGACAATACATTAGTAGCTGTTGGTACTGAACAAGGCTATGTCAACTTGTATAATGTTGAAAATGATGATATTATCTACAAGAAGTTGTTTGACAAACAGGAAGGTAGAATTCTTTGCTGCAAGTTTAATGAAACAAGCAGCATACTTGTTACAGGGTCAGTAAATACAATTAGGGTATGGAATGTCGAGACTGGACACGCAACTACACGTATATCAGTAAGCAGACGGGGTAAAGAAGTTATTGTTTGGTCCCTTGTTACACTTGAGAACAATATTATTGCATCGGGTGATAGTTTGGGTCGACTCACACTGTGGGATGGCAATACAGGTGATGAGATTGAATCATTTATTAGTCATAAAGCAGACATCTTAACAATAGCAGTGTCTGAAGACAAAAATAATTTATTCTGCAGTGGAGTGGATCCAACCATAATACAATTTAGGAAAATTAAGAAGTCTGACTCTGCAGATGAGCAATGGGTTAAGAGTTATCAAAATTCCATAAATCAGCATGATGTAAGAGATATTGCAGTATTGGATCAAAAAGTATTGTCGGTTGGTGTTGATGGATACATCTCAACATTTCATTATGTTCGAAAAGGATTTCAACGAATACCACATTTGTTGCCAACACCCAGATCATTGGTGTGTTCTTCCAAGAAATTTTTACTTTTAAGGTACAGTAATCATTTAGAAGTTTGGAAACTGGGAGCATTTCTAAAAGATAAATCAGGAAGGCCTGTAGCAGAACCTGCAAATGTAATCATTAAAAACAAAATAAATACAGAAAATAATCATTCAGACTGCGAGAAAGTAAATGAAATGTTAAAAATATCTGAAAACCCTGTGAAACTTCTATCTCTGCAGACAAAAGGAAAAAAACAGATTCAGTCATGTGCCATTTCACCAAATGGAGAGCTTATCGTTTACTCCACAGAGAGTCACACAAGGATGCTTAAATTGGATAGTGATGACGAATCGAATGTATATTTATCTAAAGTCATATTAAATGGTTCAATAACGACATGTAGCCATTTCGCTTTCACAGAAGACTCTAAAGTGTTGATCGTATATAACGCGGGTACCATCAGTTTCCTTCAAGTGGATCCTGAAGCAGGCGTCACCGTTACACAGACTATCAACACTAAAAAACATTTAAGTTGCAAATCAATATTGCATTTTCAAATATCAAAGAAATGTCCTAGCAATAAAGTGTATTTAGTGGTAGCGGACACACAAGGTGATATAGTAATATGGACACAAATTGGAAAGAAATTCGAACACTATATAACATTGCCGAAATATCATTGTGTACCATCGGCCATGGTTGTCGATGCCGAAAAGGAGACTTTGATAGTAGCATACGTTGACAGAAAGGTCGTAGAATATGAATTGTTCGAGAAAAAATTCAGTGATTGGCCCGGCTCGGCCATGCCCCCCGTGTGGTACGACACGAAGCAGTGCGTGCTGTCCGTACACCCGCACCCGAGCGGCCAGTCCGTCGTGTTCCAACACTTGAGAGATCTGTTCGTATTGGACAGGAAAGCGCAGTTTGAAATGGAGCCAACCAAAAAGAAGAAATCGGCGGACGCAGGCGAAATGAATCTAGGTTATCATATTCTTCCCATGAAGTATCTCTCCGATTTTTATTGGCTCGGCAAGAGTGAAGCTGTTACGGTCGAGATACTCCCAGAAAATATAATGTCAATGTTACCTCCCGTGGCGATAAGAAATAAACGAAGTGTTAATTAA
Protein
MATSVHRIRYYNPQPTPVNCIAYNKLNKQLSLARKDSSIEIWDMTHFPFLLKFIPAVKDGSVEALGWVKDRLLTTGLGGALVEWNLEEMCIRNTILLTGYAAWCLDVSPDNTLVAVGTEQGYVNLYNVENDDIIYKKLFDKQEGRILCCKFNETSSILVTGSVNTIRVWNVETGHATTRISVSRRGKEVIVWSLVTLENNIIASGDSLGRLTLWDGNTGDEIESFISHKADILTIAVSEDKNNLFCSGVDPTIIQFRKIKKSDSADEQWVKSYQNSINQHDVRDIAVLDQKVLSVGVDGYISTFHYVRKGFQRIPHLLPTPRSLVCSSKKFLLLRYSNHLEVWKLGAFLKDKSGRPVAEPANVIIKNKINTENNHSDCEKVNEMLKISENPVKLLSLQTKGKKQIQSCAISPNGELIVYSTESHTRMLKLDSDDESNVYLSKVILNGSITTCSHFAFTEDSKVLIVYNAGTISFLQVDPEAGVTVTQTINTKKHLSCKSILHFQISKKCPSNKVYLVVADTQGDIVIWTQIGKKFEHYITLPKYHCVPSAMVVDAEKETLIVAYVDRKVVEYELFEKKFSDWPGSAMPPVWYDTKQCVLSVHPHPSGQSVVFQHLRDLFVLDRKAQFEMEPTKKKKSADAGEMNLGYHILPMKYLSDFYWLGKSEAVTVEILPENIMSMLPPVAIRNKRSVN
Summary
Uniprot
A0A194PVC3
A0A194QNU7
A0A3S2TJC3
A0A212F414
A0A2H1VUR3
A0A0L7LRF9
+ More
A0A0L7QSZ1 A0A195ETJ6 A0A195E4R4 A0A087ZVR3 A0A2A3EAT3 A0A195B3E8 A0A154PCH4 A0A158NF52 A0A151X1K9 A0A310SGG4 A0A067R9R6 E2B6P4 F4W461 A0A2J7RKP9 A0A195CKH0 A0A026WQB7 A0A1I8M4P7 A0A1B6KAH5 A0A1B6CBI0 E9I9V9 A0A1I8PX43 A0A0A1WQI8 D6WEU4 A0A0M9A681 A0A232EKW7 K7J8T2 A0A0K8UQJ2 A0A1B0A717 A0A1A9V5V2 A0A0C9R341 A0A1Y1MM41 W8BSL2 A0A1B0C6R5 A0A1A9YSQ0 A0A182H8I2 A0A1B0G7C6 A0A0P5FVN2 A0A0P5FTQ6 A0A0P5QGW5 A0A0P5S465 A0A0N8BXE9 A0A0P5T982 A0A0P6DZF3 A0A0P4YLU4 A0A0P5SQR6 A0A0P5B794 A0A0P6FX96 A0A0P5YX22 A0A3L8D7N8 A0A0P5XW76 A0A0P5MW08 A0A182TXF9 A0A0P6C1G3 A0A0P6G6G2 A0A0N8E7I9 A0A0P5WT30 A0A0P6GCZ0 A0A0P6E1L6 A0A0P5VIU7 A0A0N8A326 A0A0P4YMC1 A0A0N8CRN3 Q7Q612 A0A0P5P412 A0A0P5U3G8 A0A0P5FSS3 A0A0P6CI28 A0A0P5QB95 A0A0P4Z4W2 A0A0P5SHL5 A0A2J7RKQ4 A0A0P5U446 A0A182XP43 A0A0N8B4W5 Q175U0 A0A0P6HSP2 A0A0N8A2S1 A0A182RGV2 A0A182I1I2 A0A0P4XLU1 B3M947 A0A0P5KT41 A0A182WHT2 A0A0P5GKK0 A0A182NMF3 A0A0P5J0F0 E9G0Z1 A0A182QTF7 A0A1Q3F5Y3 B0WXF0 A0A182ULZ9
A0A0L7QSZ1 A0A195ETJ6 A0A195E4R4 A0A087ZVR3 A0A2A3EAT3 A0A195B3E8 A0A154PCH4 A0A158NF52 A0A151X1K9 A0A310SGG4 A0A067R9R6 E2B6P4 F4W461 A0A2J7RKP9 A0A195CKH0 A0A026WQB7 A0A1I8M4P7 A0A1B6KAH5 A0A1B6CBI0 E9I9V9 A0A1I8PX43 A0A0A1WQI8 D6WEU4 A0A0M9A681 A0A232EKW7 K7J8T2 A0A0K8UQJ2 A0A1B0A717 A0A1A9V5V2 A0A0C9R341 A0A1Y1MM41 W8BSL2 A0A1B0C6R5 A0A1A9YSQ0 A0A182H8I2 A0A1B0G7C6 A0A0P5FVN2 A0A0P5FTQ6 A0A0P5QGW5 A0A0P5S465 A0A0N8BXE9 A0A0P5T982 A0A0P6DZF3 A0A0P4YLU4 A0A0P5SQR6 A0A0P5B794 A0A0P6FX96 A0A0P5YX22 A0A3L8D7N8 A0A0P5XW76 A0A0P5MW08 A0A182TXF9 A0A0P6C1G3 A0A0P6G6G2 A0A0N8E7I9 A0A0P5WT30 A0A0P6GCZ0 A0A0P6E1L6 A0A0P5VIU7 A0A0N8A326 A0A0P4YMC1 A0A0N8CRN3 Q7Q612 A0A0P5P412 A0A0P5U3G8 A0A0P5FSS3 A0A0P6CI28 A0A0P5QB95 A0A0P4Z4W2 A0A0P5SHL5 A0A2J7RKQ4 A0A0P5U446 A0A182XP43 A0A0N8B4W5 Q175U0 A0A0P6HSP2 A0A0N8A2S1 A0A182RGV2 A0A182I1I2 A0A0P4XLU1 B3M947 A0A0P5KT41 A0A182WHT2 A0A0P5GKK0 A0A182NMF3 A0A0P5J0F0 E9G0Z1 A0A182QTF7 A0A1Q3F5Y3 B0WXF0 A0A182ULZ9
Pubmed
EMBL
KQ459590
KPI97346.1
KQ461190
KPJ07198.1
RSAL01000097
RVE47670.1
+ More
AGBW02010462 OWR48474.1 ODYU01004527 SOQ44506.1 JTDY01000254 KOB78040.1 KQ414755 KOC61740.1 KQ981979 KYN31481.1 KQ979609 KYN20153.1 KZ288304 PBC28800.1 KQ976625 KYM79018.1 KQ434870 KZC09523.1 ADTU01001502 KQ982585 KYQ54287.1 KQ760208 OAD61377.1 KK852609 KDR20318.1 GL446000 EFN88628.1 GL887491 EGI71057.1 NEVH01002711 PNF41416.1 KQ977642 KYN00952.1 KK107131 EZA58210.1 GEBQ01031531 JAT08446.1 GEDC01026502 GEDC01007355 JAS10796.1 JAS29943.1 GL761846 EFZ22657.1 GBXI01013594 JAD00698.1 KQ971318 EFA00439.1 KQ435727 KOX78080.1 NNAY01003720 OXU18958.1 GDHF01023345 JAI28969.1 GBYB01001225 GBYB01001226 JAG70992.1 JAG70993.1 GEZM01027404 JAV86753.1 GAMC01006657 JAB99898.1 JXJN01026869 JXUM01029015 KQ560839 KXJ80661.1 CCAG010006832 CCAG010006833 GDIQ01253234 JAJ98490.1 GDIQ01251994 JAJ99730.1 GDIQ01135468 JAL16258.1 GDIQ01093281 JAL58445.1 GDIQ01135467 JAL16259.1 GDIP01133437 JAL70277.1 GDIQ01070739 JAN23998.1 GDIP01225471 JAI97930.1 GDIP01141739 GDIP01105585 GDIP01105582 LRGB01001581 JAL61975.1 KZS11276.1 GDIP01192924 JAJ30478.1 GDIQ01052415 JAN42322.1 GDIP01065398 JAM38317.1 QOIP01000012 RLU16266.1 GDIP01066842 JAM36873.1 GDIQ01154620 JAK97105.1 GDIP01022564 JAM81151.1 GDIQ01039306 JAN55431.1 GDIQ01054156 JAN40581.1 GDIP01081834 JAM21881.1 GDIQ01035026 JAN59711.1 GDIQ01069109 JAN25628.1 GDIP01114340 JAL89374.1 GDIP01187329 JAJ36073.1 GDIP01225470 JAI97931.1 GDIP01111829 GDIP01105583 JAL98131.1 AAAB01008960 EAA11688.4 GDIQ01135466 JAL16260.1 GDIP01118387 JAL85327.1 GDIQ01250542 JAK01183.1 GDIQ01090880 JAN03857.1 GDIQ01138128 JAL13598.1 GDIP01218525 JAJ04877.1 GDIQ01093280 JAL58446.1 PNF41417.1 GDIP01118386 JAL85328.1 GDIQ01212539 JAK39186.1 CH477395 EAT41865.1 GDIQ01014744 JAN79993.1 GDIP01188169 JAJ35233.1 APCN01000061 GDIP01241127 JAI82274.1 CH902618 EDV40031.1 GDIQ01205456 JAK46269.1 GDIQ01246576 JAK05149.1 GDIQ01208154 JAK43571.1 GL732528 EFX86991.1 AXCN02000928 GFDL01012075 JAV22970.1 DS232163 EDS36480.1
AGBW02010462 OWR48474.1 ODYU01004527 SOQ44506.1 JTDY01000254 KOB78040.1 KQ414755 KOC61740.1 KQ981979 KYN31481.1 KQ979609 KYN20153.1 KZ288304 PBC28800.1 KQ976625 KYM79018.1 KQ434870 KZC09523.1 ADTU01001502 KQ982585 KYQ54287.1 KQ760208 OAD61377.1 KK852609 KDR20318.1 GL446000 EFN88628.1 GL887491 EGI71057.1 NEVH01002711 PNF41416.1 KQ977642 KYN00952.1 KK107131 EZA58210.1 GEBQ01031531 JAT08446.1 GEDC01026502 GEDC01007355 JAS10796.1 JAS29943.1 GL761846 EFZ22657.1 GBXI01013594 JAD00698.1 KQ971318 EFA00439.1 KQ435727 KOX78080.1 NNAY01003720 OXU18958.1 GDHF01023345 JAI28969.1 GBYB01001225 GBYB01001226 JAG70992.1 JAG70993.1 GEZM01027404 JAV86753.1 GAMC01006657 JAB99898.1 JXJN01026869 JXUM01029015 KQ560839 KXJ80661.1 CCAG010006832 CCAG010006833 GDIQ01253234 JAJ98490.1 GDIQ01251994 JAJ99730.1 GDIQ01135468 JAL16258.1 GDIQ01093281 JAL58445.1 GDIQ01135467 JAL16259.1 GDIP01133437 JAL70277.1 GDIQ01070739 JAN23998.1 GDIP01225471 JAI97930.1 GDIP01141739 GDIP01105585 GDIP01105582 LRGB01001581 JAL61975.1 KZS11276.1 GDIP01192924 JAJ30478.1 GDIQ01052415 JAN42322.1 GDIP01065398 JAM38317.1 QOIP01000012 RLU16266.1 GDIP01066842 JAM36873.1 GDIQ01154620 JAK97105.1 GDIP01022564 JAM81151.1 GDIQ01039306 JAN55431.1 GDIQ01054156 JAN40581.1 GDIP01081834 JAM21881.1 GDIQ01035026 JAN59711.1 GDIQ01069109 JAN25628.1 GDIP01114340 JAL89374.1 GDIP01187329 JAJ36073.1 GDIP01225470 JAI97931.1 GDIP01111829 GDIP01105583 JAL98131.1 AAAB01008960 EAA11688.4 GDIQ01135466 JAL16260.1 GDIP01118387 JAL85327.1 GDIQ01250542 JAK01183.1 GDIQ01090880 JAN03857.1 GDIQ01138128 JAL13598.1 GDIP01218525 JAJ04877.1 GDIQ01093280 JAL58446.1 PNF41417.1 GDIP01118386 JAL85328.1 GDIQ01212539 JAK39186.1 CH477395 EAT41865.1 GDIQ01014744 JAN79993.1 GDIP01188169 JAJ35233.1 APCN01000061 GDIP01241127 JAI82274.1 CH902618 EDV40031.1 GDIQ01205456 JAK46269.1 GDIQ01246576 JAK05149.1 GDIQ01208154 JAK43571.1 GL732528 EFX86991.1 AXCN02000928 GFDL01012075 JAV22970.1 DS232163 EDS36480.1
Proteomes
UP000053268
UP000053240
UP000283053
UP000007151
UP000037510
UP000053825
+ More
UP000078541 UP000078492 UP000005203 UP000242457 UP000078540 UP000076502 UP000005205 UP000075809 UP000027135 UP000008237 UP000007755 UP000235965 UP000078542 UP000053097 UP000095301 UP000095300 UP000007266 UP000053105 UP000215335 UP000002358 UP000092445 UP000078200 UP000092460 UP000092443 UP000069940 UP000249989 UP000092444 UP000076858 UP000279307 UP000075902 UP000007062 UP000076407 UP000008820 UP000075900 UP000075840 UP000007801 UP000075920 UP000075884 UP000000305 UP000075886 UP000002320 UP000075903
UP000078541 UP000078492 UP000005203 UP000242457 UP000078540 UP000076502 UP000005205 UP000075809 UP000027135 UP000008237 UP000007755 UP000235965 UP000078542 UP000053097 UP000095301 UP000095300 UP000007266 UP000053105 UP000215335 UP000002358 UP000092445 UP000078200 UP000092460 UP000092443 UP000069940 UP000249989 UP000092444 UP000076858 UP000279307 UP000075902 UP000007062 UP000076407 UP000008820 UP000075900 UP000075840 UP000007801 UP000075920 UP000075884 UP000000305 UP000075886 UP000002320 UP000075903
Pfam
Interpro
IPR036322
WD40_repeat_dom_sf
+ More
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR024977 Apc4_WD40_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR011044 Quino_amine_DH_bsu
IPR011048 Haem_d1_sf
IPR036883 PDCD5-like_sf
IPR006624 Beta-propeller_rpt_TECPR
IPR013320 ConA-like_dom_sf
IPR002836 PDCD5-like
IPR006614 Peroxin/Ferlin
IPR018787 DUF2371_TMEM200
IPR001079 Galectin_CRD
IPR010482 Peroxin
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019775 WD40_repeat_CS
IPR024977 Apc4_WD40_dom
IPR011047 Quinoprotein_ADH-like_supfam
IPR011044 Quino_amine_DH_bsu
IPR011048 Haem_d1_sf
IPR036883 PDCD5-like_sf
IPR006624 Beta-propeller_rpt_TECPR
IPR013320 ConA-like_dom_sf
IPR002836 PDCD5-like
IPR006614 Peroxin/Ferlin
IPR018787 DUF2371_TMEM200
IPR001079 Galectin_CRD
IPR010482 Peroxin
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A194PVC3
A0A194QNU7
A0A3S2TJC3
A0A212F414
A0A2H1VUR3
A0A0L7LRF9
+ More
A0A0L7QSZ1 A0A195ETJ6 A0A195E4R4 A0A087ZVR3 A0A2A3EAT3 A0A195B3E8 A0A154PCH4 A0A158NF52 A0A151X1K9 A0A310SGG4 A0A067R9R6 E2B6P4 F4W461 A0A2J7RKP9 A0A195CKH0 A0A026WQB7 A0A1I8M4P7 A0A1B6KAH5 A0A1B6CBI0 E9I9V9 A0A1I8PX43 A0A0A1WQI8 D6WEU4 A0A0M9A681 A0A232EKW7 K7J8T2 A0A0K8UQJ2 A0A1B0A717 A0A1A9V5V2 A0A0C9R341 A0A1Y1MM41 W8BSL2 A0A1B0C6R5 A0A1A9YSQ0 A0A182H8I2 A0A1B0G7C6 A0A0P5FVN2 A0A0P5FTQ6 A0A0P5QGW5 A0A0P5S465 A0A0N8BXE9 A0A0P5T982 A0A0P6DZF3 A0A0P4YLU4 A0A0P5SQR6 A0A0P5B794 A0A0P6FX96 A0A0P5YX22 A0A3L8D7N8 A0A0P5XW76 A0A0P5MW08 A0A182TXF9 A0A0P6C1G3 A0A0P6G6G2 A0A0N8E7I9 A0A0P5WT30 A0A0P6GCZ0 A0A0P6E1L6 A0A0P5VIU7 A0A0N8A326 A0A0P4YMC1 A0A0N8CRN3 Q7Q612 A0A0P5P412 A0A0P5U3G8 A0A0P5FSS3 A0A0P6CI28 A0A0P5QB95 A0A0P4Z4W2 A0A0P5SHL5 A0A2J7RKQ4 A0A0P5U446 A0A182XP43 A0A0N8B4W5 Q175U0 A0A0P6HSP2 A0A0N8A2S1 A0A182RGV2 A0A182I1I2 A0A0P4XLU1 B3M947 A0A0P5KT41 A0A182WHT2 A0A0P5GKK0 A0A182NMF3 A0A0P5J0F0 E9G0Z1 A0A182QTF7 A0A1Q3F5Y3 B0WXF0 A0A182ULZ9
A0A0L7QSZ1 A0A195ETJ6 A0A195E4R4 A0A087ZVR3 A0A2A3EAT3 A0A195B3E8 A0A154PCH4 A0A158NF52 A0A151X1K9 A0A310SGG4 A0A067R9R6 E2B6P4 F4W461 A0A2J7RKP9 A0A195CKH0 A0A026WQB7 A0A1I8M4P7 A0A1B6KAH5 A0A1B6CBI0 E9I9V9 A0A1I8PX43 A0A0A1WQI8 D6WEU4 A0A0M9A681 A0A232EKW7 K7J8T2 A0A0K8UQJ2 A0A1B0A717 A0A1A9V5V2 A0A0C9R341 A0A1Y1MM41 W8BSL2 A0A1B0C6R5 A0A1A9YSQ0 A0A182H8I2 A0A1B0G7C6 A0A0P5FVN2 A0A0P5FTQ6 A0A0P5QGW5 A0A0P5S465 A0A0N8BXE9 A0A0P5T982 A0A0P6DZF3 A0A0P4YLU4 A0A0P5SQR6 A0A0P5B794 A0A0P6FX96 A0A0P5YX22 A0A3L8D7N8 A0A0P5XW76 A0A0P5MW08 A0A182TXF9 A0A0P6C1G3 A0A0P6G6G2 A0A0N8E7I9 A0A0P5WT30 A0A0P6GCZ0 A0A0P6E1L6 A0A0P5VIU7 A0A0N8A326 A0A0P4YMC1 A0A0N8CRN3 Q7Q612 A0A0P5P412 A0A0P5U3G8 A0A0P5FSS3 A0A0P6CI28 A0A0P5QB95 A0A0P4Z4W2 A0A0P5SHL5 A0A2J7RKQ4 A0A0P5U446 A0A182XP43 A0A0N8B4W5 Q175U0 A0A0P6HSP2 A0A0N8A2S1 A0A182RGV2 A0A182I1I2 A0A0P4XLU1 B3M947 A0A0P5KT41 A0A182WHT2 A0A0P5GKK0 A0A182NMF3 A0A0P5J0F0 E9G0Z1 A0A182QTF7 A0A1Q3F5Y3 B0WXF0 A0A182ULZ9
PDB
5N1A
E-value=8.91596e-35,
Score=370
Ontologies
GO
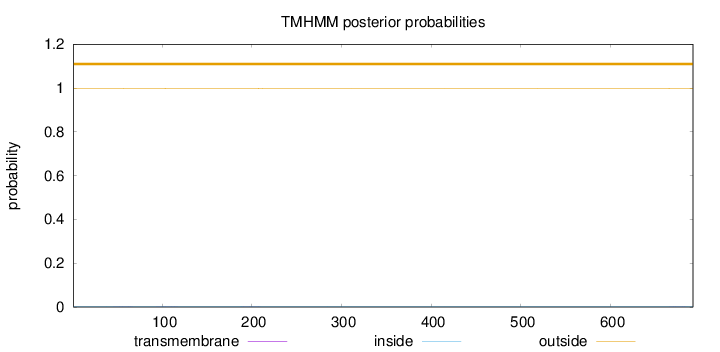
Topology
Length:
692
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03121
Exp number, first 60 AAs:
0.00326
Total prob of N-in:
0.00268
outside
1 - 692
Population Genetic Test Statistics
Pi
222.461954
Theta
150.61613
Tajima's D
-1.197855
CLR
154.949307
CSRT
0.105194740262987
Interpretation
Uncertain
