Gene
KWMTBOMO06604
Pre Gene Modal
BGIBMGA011797
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_DNA_excision_repair_protein_haywire_[Bombyx_mori]
Full name
General transcription and DNA repair factor IIH helicase subunit XPB
Alternative Name
ATP-dependent DNA helicase hay
DNA excision repair protein haywire
ERCC-3 homolog protein
ERCC3Dm
DNA excision repair protein haywire
ERCC-3 homolog protein
ERCC3Dm
Location in the cell
Nuclear Reliability : 2.616
Sequence
CDS
ATGGGGCCCCCGAAGAAATTCAAGAGATATGACTCAAAAGGAGGAAGTGATAGATCAAGTAAAAAGAAAAAAGTAGATGAGGATATAACAATAGATCTTGTAGATGATGATAATGCAGAAAACGATGGGGTCCCTGTTGCTGCCATGCAAGATGCTGAGAAGAATGATCAAGTACCTGAAGACGAATTTGGTGCTAAAGATTATAGAAGTCAAATGCCGCTTAAACCAGACAACGCCAGTCGTCCTTTATGGGTGGCACCCAATGGACATATTTTCTTGGAAGCATTTTCTCCAGTTTACAAACATGCTCATGATTTTTTGATTGCTATAGCTGAACCAGTTTGCAGACCCCAACACATACATGAATACAAATTGACAGCGTACAGTTTGTACGCGGCAGTTTCGGTGGGCCTACAGACCAATGACATTATTGAGTACCTGCAGAGACTGAGCAAGTGTGCTGTACCGGCCGGCATCGTAGAGTTCATTAAACTATGTACCCTCTCTTATGGTAAAGTGAAGCTGGTGTTAAAACATAACAGGTATCTAGTCGAGAGTAAGCACGTGGAGGTACTGCAGAAGCTATTGAAAGATCCAGTGATTCAACAGTGCAGGCTGCGACGCGACGGCGACGAGGATCTGCTGTCCAACGACCTGCCCGCCAAGCCAGCCACGTCCCTCGCCAAACCAGGTGAAGAAAAACCACCGGCGGCGAACGGCGCTGTGCCCGACGACATCAGTCAGTTCTATCAGCAACTAGATAAGGATGACGACGAGGAGGAGACCTCTGGTGTTACTGCCAACGCAGCTGTTGCCTTTGAAGTGGATCCTGACAAAATCGAAGTAATACAAAAACGTTGTATCGAGTTGGAGCATCCATTGCTGGCTGAGTACGACTTCCGGAACGACTCCATCAATCCGGACATCAACATTGATCTTAAACCAACTGCCGTTTTGCGACCTTATCAAGAGAAGAGCTTGAGGAAAATGTTCGGCAACGGCAGGGCGAGGTCAGGCGTGATAGTGTTGCCGTGCGGCGCCGGCAAGTCGCTGGTGGGCGTGACGGCGGTGTGCACGGTGCGCAAGCGCGCCCTCGTGCTCTGCAACTCCGGCGTCTCCGTGGAACAGTGGAAGCAACAGTTCAAGTGCTGGTCCACGGCGGACGACAGCATGATCTGCAGGTTCACATCAGAAGCCAAGGACAAGCCGATGGGAGCGGGTATCCTCGTAACGACGTATTCCATGATAACTCACGGACAGAGACGCTCGTGGGAGGCCGAACAGACGATGAAGTGGCTCCAGGCCCAGGAGTGGGGCCTGGTCGTGCTCGACGAAGTGCACACGATACCTGCCAAAATGTTCCGCAGGGTGCTCACCATCGTGCACTCGCACGCCAAACTGGGTTTGACGGCGACCCTGCTGCGAGAGGACGACAAGATCGCGGACCTGAACTTCCTGATAGGCCCGAAGCTGTACGAGGCGAACTGGCTCGAGCTGCAGGCGGCCGGCTACATCGCGCGCGTGCAGTGCGCCGAGGTGTGGTGCCCCATGACGCCAGAGTTCTACAGGGAATACCTAGTGCAGAAAATAAACAAGAAAATGTTGTTGTACGTAATGAACCCGTCGAAGTTCCGCGCGTGCCAGTACCTGGTGCGCTACCACGAGCGGCGCGGCGACAAGACCATCGTGTTCTCCGACAACGTGTTCGCGCTCAGGCACTACGCCGTCAAGATGAACAAGCCATACATCTACGGGCCGACGTCCCAGAGCGAGCGAATACAGATCCTCCAGAATTTCAAGTTCAACCCTAAAGTGAATACGATATTCGTGAGTAAAGTCGCCGACACCAGCTTCGATCTGCCCGAAGCGAACGTGCTTATACAGATCTCTTCGCACGGAGGCTCCAGGCGACAAGAGGCACAGAGATTAGGTCGTATTTTAAGAGCAAAGAAAGGCGCTTTGGCCGAAGAATATAATGCCTTTTTCTACACTCTAGTATCACAAGACACCTTGGAGATGGCTTACAGTAGAAAAAGACAAAGATTTCTTGTTAATCAGGGTTACAGTTATAAGGTTATAACAGAACTAAAAGGTATGGATCAAGAGCCCGATATGCTGTATGGCACCCGCGAGGAACAAGGCACTTTACTCTCACAGGTGCTGGCGGCCTCTGAGACAGAGTGCGAGGAGGAGCGGGAGGGCGCGGTGGGGGGCGTGTCGCGGCGGGGGGGCGCGCTGTCGTCGCTGGCGGGCGCGGACGACGCGCTCTACGTGGAGCGCCGCGGCAACCAACACAACAAACATCCTCTCTTCAAGAAGTTCCGATATTAG
Protein
MGPPKKFKRYDSKGGSDRSSKKKKVDEDITIDLVDDDNAENDGVPVAAMQDAEKNDQVPEDEFGAKDYRSQMPLKPDNASRPLWVAPNGHIFLEAFSPVYKHAHDFLIAIAEPVCRPQHIHEYKLTAYSLYAAVSVGLQTNDIIEYLQRLSKCAVPAGIVEFIKLCTLSYGKVKLVLKHNRYLVESKHVEVLQKLLKDPVIQQCRLRRDGDEDLLSNDLPAKPATSLAKPGEEKPPAANGAVPDDISQFYQQLDKDDDEEETSGVTANAAVAFEVDPDKIEVIQKRCIELEHPLLAEYDFRNDSINPDINIDLKPTAVLRPYQEKSLRKMFGNGRARSGVIVLPCGAGKSLVGVTAVCTVRKRALVLCNSGVSVEQWKQQFKCWSTADDSMICRFTSEAKDKPMGAGILVTTYSMITHGQRRSWEAEQTMKWLQAQEWGLVVLDEVHTIPAKMFRRVLTIVHSHAKLGLTATLLREDDKIADLNFLIGPKLYEANWLELQAAGYIARVQCAEVWCPMTPEFYREYLVQKINKKMLLYVMNPSKFRACQYLVRYHERRGDKTIVFSDNVFALRHYAVKMNKPYIYGPTSQSERIQILQNFKFNPKVNTIFVSKVADTSFDLPEANVLIQISSHGGSRRQEAQRLGRILRAKKGALAEEYNAFFYTLVSQDTLEMAYSRKRQRFLVNQGYSYKVITELKGMDQEPDMLYGTREEQGTLLSQVLAASETECEEEREGAVGGVSRRGGALSSLAGADDALYVERRGNQHNKHPLFKKFRY
Summary
Description
ATP-dependent 3'-5' DNA helicase, component of the general transcription and DNA repair factor IIH (TFIIH) core complex, which is involved in general and transcription-coupled nucleotide excision repair (NER) of damaged DNA and, when complexed to CAK, in RNA transcription by RNA polymerase II. In NER, TFIIH acts by opening DNA around the lesion to allow the excision of the damaged oligonucleotide and its replacement by a new DNA fragment. The ATPase activity of haywire/XPB/ERCC3, but not its helicase activity, is required for DNA opening. In transcription, TFIIH has an essential role in transcription initiation. When the pre-initiation complex (PIC) has been established, TFIIH is required for promoter opening and promoter escape. The ATP-dependent helicase activity of haywire/XPB/ERCC3 is required for promoter opening and promoter escape. Phosphorylation of the C-terminal tail (CTD) of the largest subunit of RNA polymerase II by the kinase module CAK controls the initiation of transcription.
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Subunit
Component of the 7-subunit TFIIH core complex composed of haywire/XPB/ERCC3, XPD/ERCC2, GTF2H1, GTF2H2, GTF2H3, GTF2H4 and GTF2H5, which is active in NER. The core complex associates with the 3-subunit CDK-activating kinase (CAK) module composed of CCNH/cyclin H, CDK7 and MNAT1 to form the 10-subunit holoenzyme (holo-TFIIH) active in transcription. Interacts with PUF60. Interacts with ATF7IP. Interacts with Epstein-Barr virus EBNA2.
Similarity
Belongs to the helicase family. RAD25/XPB subfamily.
Keywords
ATP-binding
Complete proteome
DNA damage
DNA repair
DNA-binding
Helicase
Hydrolase
Nucleotide-binding
Nucleus
Reference proteome
Feature
chain General transcription and DNA repair factor IIH helicase subunit XPB
Uniprot
A0A3S2LKI1
A0A2W1BHW1
A0A194QQG3
A0A194PX48
D6WEU3
A0A336M8X8
+ More
A0A336M4N2 N6TWJ3 N6TZ62 U4UBP8 A0A0T6BCJ9 A0A1Q3F426 A0A1B6E978 Q16KH8 Q16JU4 B0WI20 A0A182H9N0 W8BI97 A0A084WUJ8 A0A1W5C8V3 A0A0K8UYD2 A0A1S4HDZ5 A0A182IZP8 A0A182HPS5 A0A182VML4 A0A182PGC3 A0A182UJ71 A0A182MXB9 A0A182X003 A0A1I8MEY2 W5JXB0 T1PJB3 A0A182QX29 A0A1I8QDV0 A0A154P0R8 A0A026WFU6 A0A182WD96 A0A182FW78 A0A182RAJ0 A0A0L0BR20 A0A0A1XNT6 A0A088A138 A0A182MQ73 A0A1B6GA12 B4J1K5 A0A2A3EN14 B4HLV8 B4N6Q5 B3NCL7 A0A1B0CNG7 A0A0L7QUH7 B4PE26 B7Z0G1 Q02870 B4KVX7 B4LET7 A0A0P4VST7 F4WR81 A0A1Y1MEN7 A0A1W4UWP2 B4QP78 A0A0V0G770 A0A158NVF5 A0A158NVF4 A0A069DWY2 A0A0J9RSS4 E0VCI8 A0A232F865 A0A1Q3F4C1 A0A151J2Z8 A0A224X7U7 A0A023F5L2 A0A2R7WGC3 A0A0M3QWA1 A0A067QZB7 B3M5C1 Q2LZJ4 B4H6P5 A0A1A9VYG8 E2AW10 A0A1A9Z3I2 A0A0M9A381 A0A3B0J514 A0A1A9Y9J3 A0A1A9WU53 A0A1L8DGX6 A0A1J1HXV6 A0A1B0FC49 A0A2Z5TU28 A0A151X0B3 A0A146LTX2 A0A151I3R9 T1HPF2 K7J022 A0A182YQ89 L7MF25 A0A1E1X4Q9 A0A131XN85 A0A023FMB8 A0A224YRL5
A0A336M4N2 N6TWJ3 N6TZ62 U4UBP8 A0A0T6BCJ9 A0A1Q3F426 A0A1B6E978 Q16KH8 Q16JU4 B0WI20 A0A182H9N0 W8BI97 A0A084WUJ8 A0A1W5C8V3 A0A0K8UYD2 A0A1S4HDZ5 A0A182IZP8 A0A182HPS5 A0A182VML4 A0A182PGC3 A0A182UJ71 A0A182MXB9 A0A182X003 A0A1I8MEY2 W5JXB0 T1PJB3 A0A182QX29 A0A1I8QDV0 A0A154P0R8 A0A026WFU6 A0A182WD96 A0A182FW78 A0A182RAJ0 A0A0L0BR20 A0A0A1XNT6 A0A088A138 A0A182MQ73 A0A1B6GA12 B4J1K5 A0A2A3EN14 B4HLV8 B4N6Q5 B3NCL7 A0A1B0CNG7 A0A0L7QUH7 B4PE26 B7Z0G1 Q02870 B4KVX7 B4LET7 A0A0P4VST7 F4WR81 A0A1Y1MEN7 A0A1W4UWP2 B4QP78 A0A0V0G770 A0A158NVF5 A0A158NVF4 A0A069DWY2 A0A0J9RSS4 E0VCI8 A0A232F865 A0A1Q3F4C1 A0A151J2Z8 A0A224X7U7 A0A023F5L2 A0A2R7WGC3 A0A0M3QWA1 A0A067QZB7 B3M5C1 Q2LZJ4 B4H6P5 A0A1A9VYG8 E2AW10 A0A1A9Z3I2 A0A0M9A381 A0A3B0J514 A0A1A9Y9J3 A0A1A9WU53 A0A1L8DGX6 A0A1J1HXV6 A0A1B0FC49 A0A2Z5TU28 A0A151X0B3 A0A146LTX2 A0A151I3R9 T1HPF2 K7J022 A0A182YQ89 L7MF25 A0A1E1X4Q9 A0A131XN85 A0A023FMB8 A0A224YRL5
EC Number
3.6.4.12
Pubmed
28756777
26354079
18362917
19820115
23537049
17510324
+ More
26483478 24495485 24438588 12364791 25315136 20920257 23761445 24508170 26108605 25830018 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1454518 1458540 12537569 27129103 21719571 28004739 21347285 26334808 22936249 20566863 28648823 25474469 24845553 15632085 20798317 26760975 26823975 20075255 25244985 25576852 28503490 28049606 28797301
26483478 24495485 24438588 12364791 25315136 20920257 23761445 24508170 26108605 25830018 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1454518 1458540 12537569 27129103 21719571 28004739 21347285 26334808 22936249 20566863 28648823 25474469 24845553 15632085 20798317 26760975 26823975 20075255 25244985 25576852 28503490 28049606 28797301
EMBL
RSAL01000080
RVE48630.1
KZ150049
PZC74438.1
KQ461190
KPJ07205.1
+ More
KQ459590 KPI97339.1 KQ971318 EFA00328.1 UFQT01000554 SSX25233.1 SSX25232.1 APGK01049768 APGK01049769 KB741156 ENN73635.1 ENN73636.1 KB631941 ERL87360.1 LJIG01001860 KRT85080.1 GFDL01012758 JAV22287.1 GEDC01002806 JAS34492.1 CH477957 EAT34802.1 CH477990 EAT34570.1 DS231941 EDS28102.1 JXUM01121146 KQ566467 KXJ70112.1 GAMC01017131 GAMC01017129 JAB89424.1 ATLV01027094 KE525423 KFB53892.1 AAAB01008986 EAA00171.4 GDHF01020761 JAI31553.1 APCN01003270 ADMH02000039 ETN68024.1 KA648210 AFP62839.1 AXCN02001682 KQ434792 KZC05451.1 KK107234 EZA54927.1 JRES01001499 KNC22428.1 GBXI01001631 JAD12661.1 AXCM01007405 GECZ01010507 JAS59262.1 CH916366 EDV96925.1 KZ288215 PBC32652.1 CH480815 EDW40990.1 CH964161 EDW80044.1 CH954178 EDV51314.1 AJWK01020298 AJWK01020299 KQ414735 KOC62214.1 CM000159 EDW94022.1 AE014296 ACL83286.1 S50517 X68309 L02965 AY051975 BT025199 CH933809 EDW19528.2 CH940647 EDW70194.1 GDKW01001034 JAI55561.1 GL888284 EGI63247.1 GEZM01033278 JAV84289.1 CM000363 EDX09976.1 GECL01002564 JAP03560.1 ADTU01027132 GBGD01000553 JAC88336.1 CM002912 KMY98856.1 DS235053 EEB11094.1 NNAY01000790 OXU26497.1 GFDL01012633 JAV22412.1 KQ980313 KYN16701.1 GFTR01008001 JAW08425.1 GBBI01002263 JAC16449.1 KK854787 PTY18714.1 CP012525 ALC43783.1 KK853163 KDR10417.1 CH902618 EDV40626.1 CH379069 EAL29513.2 CH479214 EDW33473.1 GL443213 EFN62432.1 KQ435776 KOX74919.1 OUUW01000002 SPP76695.1 GFDF01008490 JAV05594.1 CVRI01000036 CRK92919.1 CCAG010016063 FX985847 BBA93734.1 KQ982617 KYQ53772.1 GDHC01011125 GDHC01007930 JAQ07504.1 JAQ10699.1 KQ976478 KYM83804.1 ACPB03017115 ACPB03017116 ACPB03017117 GACK01003245 JAA61789.1 GFAC01004944 JAT94244.1 GEFH01001560 JAP67021.1 GBBK01002103 JAC22379.1 GFPF01007253 MAA18399.1
KQ459590 KPI97339.1 KQ971318 EFA00328.1 UFQT01000554 SSX25233.1 SSX25232.1 APGK01049768 APGK01049769 KB741156 ENN73635.1 ENN73636.1 KB631941 ERL87360.1 LJIG01001860 KRT85080.1 GFDL01012758 JAV22287.1 GEDC01002806 JAS34492.1 CH477957 EAT34802.1 CH477990 EAT34570.1 DS231941 EDS28102.1 JXUM01121146 KQ566467 KXJ70112.1 GAMC01017131 GAMC01017129 JAB89424.1 ATLV01027094 KE525423 KFB53892.1 AAAB01008986 EAA00171.4 GDHF01020761 JAI31553.1 APCN01003270 ADMH02000039 ETN68024.1 KA648210 AFP62839.1 AXCN02001682 KQ434792 KZC05451.1 KK107234 EZA54927.1 JRES01001499 KNC22428.1 GBXI01001631 JAD12661.1 AXCM01007405 GECZ01010507 JAS59262.1 CH916366 EDV96925.1 KZ288215 PBC32652.1 CH480815 EDW40990.1 CH964161 EDW80044.1 CH954178 EDV51314.1 AJWK01020298 AJWK01020299 KQ414735 KOC62214.1 CM000159 EDW94022.1 AE014296 ACL83286.1 S50517 X68309 L02965 AY051975 BT025199 CH933809 EDW19528.2 CH940647 EDW70194.1 GDKW01001034 JAI55561.1 GL888284 EGI63247.1 GEZM01033278 JAV84289.1 CM000363 EDX09976.1 GECL01002564 JAP03560.1 ADTU01027132 GBGD01000553 JAC88336.1 CM002912 KMY98856.1 DS235053 EEB11094.1 NNAY01000790 OXU26497.1 GFDL01012633 JAV22412.1 KQ980313 KYN16701.1 GFTR01008001 JAW08425.1 GBBI01002263 JAC16449.1 KK854787 PTY18714.1 CP012525 ALC43783.1 KK853163 KDR10417.1 CH902618 EDV40626.1 CH379069 EAL29513.2 CH479214 EDW33473.1 GL443213 EFN62432.1 KQ435776 KOX74919.1 OUUW01000002 SPP76695.1 GFDF01008490 JAV05594.1 CVRI01000036 CRK92919.1 CCAG010016063 FX985847 BBA93734.1 KQ982617 KYQ53772.1 GDHC01011125 GDHC01007930 JAQ07504.1 JAQ10699.1 KQ976478 KYM83804.1 ACPB03017115 ACPB03017116 ACPB03017117 GACK01003245 JAA61789.1 GFAC01004944 JAT94244.1 GEFH01001560 JAP67021.1 GBBK01002103 JAC22379.1 GFPF01007253 MAA18399.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000007266
UP000019118
UP000030742
+ More
UP000008820 UP000002320 UP000069940 UP000249989 UP000030765 UP000007062 UP000075880 UP000075840 UP000075903 UP000075885 UP000075902 UP000075884 UP000076407 UP000095301 UP000000673 UP000075886 UP000095300 UP000076502 UP000053097 UP000075920 UP000069272 UP000075900 UP000037069 UP000005203 UP000075883 UP000001070 UP000242457 UP000001292 UP000007798 UP000008711 UP000092461 UP000053825 UP000002282 UP000000803 UP000009192 UP000008792 UP000007755 UP000192221 UP000000304 UP000005205 UP000009046 UP000215335 UP000078492 UP000092553 UP000027135 UP000007801 UP000001819 UP000008744 UP000078200 UP000000311 UP000092445 UP000053105 UP000268350 UP000092443 UP000091820 UP000183832 UP000092444 UP000075809 UP000078540 UP000015103 UP000002358 UP000076408
UP000008820 UP000002320 UP000069940 UP000249989 UP000030765 UP000007062 UP000075880 UP000075840 UP000075903 UP000075885 UP000075902 UP000075884 UP000076407 UP000095301 UP000000673 UP000075886 UP000095300 UP000076502 UP000053097 UP000075920 UP000069272 UP000075900 UP000037069 UP000005203 UP000075883 UP000001070 UP000242457 UP000001292 UP000007798 UP000008711 UP000092461 UP000053825 UP000002282 UP000000803 UP000009192 UP000008792 UP000007755 UP000192221 UP000000304 UP000005205 UP000009046 UP000215335 UP000078492 UP000092553 UP000027135 UP000007801 UP000001819 UP000008744 UP000078200 UP000000311 UP000092445 UP000053105 UP000268350 UP000092443 UP000091820 UP000183832 UP000092444 UP000075809 UP000078540 UP000015103 UP000002358 UP000076408
Pfam
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A3S2LKI1
A0A2W1BHW1
A0A194QQG3
A0A194PX48
D6WEU3
A0A336M8X8
+ More
A0A336M4N2 N6TWJ3 N6TZ62 U4UBP8 A0A0T6BCJ9 A0A1Q3F426 A0A1B6E978 Q16KH8 Q16JU4 B0WI20 A0A182H9N0 W8BI97 A0A084WUJ8 A0A1W5C8V3 A0A0K8UYD2 A0A1S4HDZ5 A0A182IZP8 A0A182HPS5 A0A182VML4 A0A182PGC3 A0A182UJ71 A0A182MXB9 A0A182X003 A0A1I8MEY2 W5JXB0 T1PJB3 A0A182QX29 A0A1I8QDV0 A0A154P0R8 A0A026WFU6 A0A182WD96 A0A182FW78 A0A182RAJ0 A0A0L0BR20 A0A0A1XNT6 A0A088A138 A0A182MQ73 A0A1B6GA12 B4J1K5 A0A2A3EN14 B4HLV8 B4N6Q5 B3NCL7 A0A1B0CNG7 A0A0L7QUH7 B4PE26 B7Z0G1 Q02870 B4KVX7 B4LET7 A0A0P4VST7 F4WR81 A0A1Y1MEN7 A0A1W4UWP2 B4QP78 A0A0V0G770 A0A158NVF5 A0A158NVF4 A0A069DWY2 A0A0J9RSS4 E0VCI8 A0A232F865 A0A1Q3F4C1 A0A151J2Z8 A0A224X7U7 A0A023F5L2 A0A2R7WGC3 A0A0M3QWA1 A0A067QZB7 B3M5C1 Q2LZJ4 B4H6P5 A0A1A9VYG8 E2AW10 A0A1A9Z3I2 A0A0M9A381 A0A3B0J514 A0A1A9Y9J3 A0A1A9WU53 A0A1L8DGX6 A0A1J1HXV6 A0A1B0FC49 A0A2Z5TU28 A0A151X0B3 A0A146LTX2 A0A151I3R9 T1HPF2 K7J022 A0A182YQ89 L7MF25 A0A1E1X4Q9 A0A131XN85 A0A023FMB8 A0A224YRL5
A0A336M4N2 N6TWJ3 N6TZ62 U4UBP8 A0A0T6BCJ9 A0A1Q3F426 A0A1B6E978 Q16KH8 Q16JU4 B0WI20 A0A182H9N0 W8BI97 A0A084WUJ8 A0A1W5C8V3 A0A0K8UYD2 A0A1S4HDZ5 A0A182IZP8 A0A182HPS5 A0A182VML4 A0A182PGC3 A0A182UJ71 A0A182MXB9 A0A182X003 A0A1I8MEY2 W5JXB0 T1PJB3 A0A182QX29 A0A1I8QDV0 A0A154P0R8 A0A026WFU6 A0A182WD96 A0A182FW78 A0A182RAJ0 A0A0L0BR20 A0A0A1XNT6 A0A088A138 A0A182MQ73 A0A1B6GA12 B4J1K5 A0A2A3EN14 B4HLV8 B4N6Q5 B3NCL7 A0A1B0CNG7 A0A0L7QUH7 B4PE26 B7Z0G1 Q02870 B4KVX7 B4LET7 A0A0P4VST7 F4WR81 A0A1Y1MEN7 A0A1W4UWP2 B4QP78 A0A0V0G770 A0A158NVF5 A0A158NVF4 A0A069DWY2 A0A0J9RSS4 E0VCI8 A0A232F865 A0A1Q3F4C1 A0A151J2Z8 A0A224X7U7 A0A023F5L2 A0A2R7WGC3 A0A0M3QWA1 A0A067QZB7 B3M5C1 Q2LZJ4 B4H6P5 A0A1A9VYG8 E2AW10 A0A1A9Z3I2 A0A0M9A381 A0A3B0J514 A0A1A9Y9J3 A0A1A9WU53 A0A1L8DGX6 A0A1J1HXV6 A0A1B0FC49 A0A2Z5TU28 A0A151X0B3 A0A146LTX2 A0A151I3R9 T1HPF2 K7J022 A0A182YQ89 L7MF25 A0A1E1X4Q9 A0A131XN85 A0A023FMB8 A0A224YRL5
PDB
6O9M
E-value=0,
Score=2634
Ontologies
PATHWAY
GO
GO:0004003
GO:0003677
GO:0007064
GO:0031390
GO:0006367
GO:0006289
GO:0005524
GO:0043138
GO:0006468
GO:0033683
GO:0006366
GO:0097550
GO:0009411
GO:0005675
GO:0008134
GO:0000112
GO:0005525
GO:0007264
GO:0003924
GO:0040008
GO:1903025
GO:0042127
GO:0006355
GO:0000381
GO:0043140
GO:0005634
GO:0001111
GO:0004386
GO:0001113
GO:0016787
GO:0006811
GO:0016020
GO:0006260
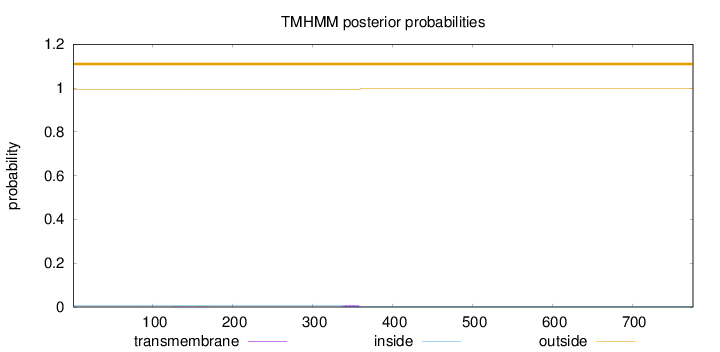
Topology
Subcellular location
Nucleus
Length:
776
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19172
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00728
outside
1 - 776
Population Genetic Test Statistics
Pi
175.578871
Theta
155.025869
Tajima's D
0.09552
CLR
211.003982
CSRT
0.400029998500075
Interpretation
Uncertain
