Gene
KWMTBOMO06602
Pre Gene Modal
BGIBMGA011798
Annotation
Leukocyte_receptor_cluster_member_1_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 3.526
Sequence
CDS
ATGAATATCTTGCCAAAAAAAAGGTGGCATGTTCGGACTAAGGAAAATATTGCAAGGGTTAGAAAGGATGAAGCAGAAGCTGCAGAGAAGGAAAAACAAGAAAGAATAAGAACAGAAACTGCAGATAAAGAGTCAAGGCTCAATATACTAAAGCAAAAAAGCAAAGAGAGATTACTCAAATCAGGAATCCCACAAACAAATGTAGGAAAAAGTGAAATTATAAATGAACATGTCAACTTATTTGCAAATCTTGAAGATGCTATTAAAACAACAAACAAAGAACATGATAAAGAGGTAAAGGCCAAACAAGAAGAATATGAAAAACAAATTGGCTATTTAACTTATCTTGGACAAGATACAAATGAAGCTTTAAAAAAGAAAAATTGGTATGATGTTCCACCAGAGGCTTCTAGATATTCACGCACCAATGACATAAAAGATACCTATAATAAGCTAGTTTTGAAAGATGAAGATGGAAAACCAAAAACAAATTGTGTGGAGATTAAAAATGGAGAATTCGCTTGGAAACAGAAGCAACGACTAGATCCCATGAATCACATAAAAAGAAACAAAAGTTACAAACTTCAAAGTAACGAACCAAATACAAAGGTACATAAGAAAAACAAAAATCGTAATGACGAAAAGTGCCCAAAAATGAAACTACAAAAACTAAGAGAAGCTCGCCTTAAGAGAGAACAGCAGGAAAGGTGTAGAACAGAATTATTTCTAAAAAAATTAGGTGGGTTTGGTGAAAGTTCAACAGAAGAAAACAATGGAAAAAATAAACTGAATATTATTCCTAAATATAATTCTCAGTTTAACCCTGAATTGGCAAGACAGAATTATAAATGA
Protein
MNILPKKRWHVRTKENIARVRKDEAEAAEKEKQERIRTETADKESRLNILKQKSKERLLKSGIPQTNVGKSEIINEHVNLFANLEDAIKTTNKEHDKEVKAKQEEYEKQIGYLTYLGQDTNEALKKKNWYDVPPEASRYSRTNDIKDTYNKLVLKDEDGKPKTNCVEIKNGEFAWKQKQRLDPMNHIKRNKSYKLQSNEPNTKVHKKNKNRNDEKCPKMKLQKLREARLKREQQERCRTELFLKKLGGFGESSTEENNGKNKLNIIPKYNSQFNPELARQNYK
Summary
Uniprot
H9JQJ2
A0A2H1W7Y4
A0A0L7L9R1
A0A2A4JPD2
A0A194QPD1
A0A194PVI6
+ More
A0A212F3Z9 A0A3S2NZP6 A0A182PGA5 A0A1Y1KV23 A0A182V7P1 A0A195CJS7 A0A034W365 A0A151X1H8 A0A0K8V5X7 A0A1B0EYK1 A0A1I8N8N7 A0A1A9W2B1 A0A195B3Y5 A0A0A1XQZ4 Q9VH00 E2BK71 A0A0M3QY48 A0A195ETN0 A0A2H8TQS4 A0A2S2NSP7 A0A195DVF0 A0A067QGG2 B4HIU4 B4QV91 A0A154PS57 F4W432 J9K0A9 A0A1I8PJ24 A0A0J7JX82 A0A2A3E8Q5 A0A0L7QSY9 C4WWF4 A0A158NK02 L7M4D9 A0A2J7QN15 A0A131YSB0 N6T678 A0A3B0K066 J3JYI0 A0A224YGQ6 A0A3L8D7C5 A0A182IVF9 A0A088AAD8 A0A2M4CII5 A0A087UTT9 U4UKY2 A0A182JPV3 T1HPZ0 A0A0L0CBB0 E9I9T6 A0A2R7VTT8 A0A0M3JXB8 A0A182LZ25 A0A182G4Y7 U5EZW6 A0A084WDY1 A0A182HQN6 A0A0K0JMN7 A0A182H754 J9F255 B3P1P1 A0A182U7A6 Q7Q7W7 A0A182XJI1 A0A182QJ85 T1IHK6 A0A1Q3FWN0 A0A2P8YWY8 T1IJ04 A0A183D3C9 A0A0V1AH27 Q16IZ6
A0A212F3Z9 A0A3S2NZP6 A0A182PGA5 A0A1Y1KV23 A0A182V7P1 A0A195CJS7 A0A034W365 A0A151X1H8 A0A0K8V5X7 A0A1B0EYK1 A0A1I8N8N7 A0A1A9W2B1 A0A195B3Y5 A0A0A1XQZ4 Q9VH00 E2BK71 A0A0M3QY48 A0A195ETN0 A0A2H8TQS4 A0A2S2NSP7 A0A195DVF0 A0A067QGG2 B4HIU4 B4QV91 A0A154PS57 F4W432 J9K0A9 A0A1I8PJ24 A0A0J7JX82 A0A2A3E8Q5 A0A0L7QSY9 C4WWF4 A0A158NK02 L7M4D9 A0A2J7QN15 A0A131YSB0 N6T678 A0A3B0K066 J3JYI0 A0A224YGQ6 A0A3L8D7C5 A0A182IVF9 A0A088AAD8 A0A2M4CII5 A0A087UTT9 U4UKY2 A0A182JPV3 T1HPZ0 A0A0L0CBB0 E9I9T6 A0A2R7VTT8 A0A0M3JXB8 A0A182LZ25 A0A182G4Y7 U5EZW6 A0A084WDY1 A0A182HQN6 A0A0K0JMN7 A0A182H754 J9F255 B3P1P1 A0A182U7A6 Q7Q7W7 A0A182XJI1 A0A182QJ85 T1IHK6 A0A1Q3FWN0 A0A2P8YWY8 T1IJ04 A0A183D3C9 A0A0V1AH27 Q16IZ6
Pubmed
19121390
26227816
26354079
22118469
28004739
25348373
+ More
25315136 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 24845553 17994087 21719571 21347285 25576852 26830274 23537049 22516182 28797301 30249741 26108605 21282665 26483478 24438588 17885136 26850696 12364791 14747013 17210077 29403074 17510324
25315136 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 24845553 17994087 21719571 21347285 25576852 26830274 23537049 22516182 28797301 30249741 26108605 21282665 26483478 24438588 17885136 26850696 12364791 14747013 17210077 29403074 17510324
EMBL
BABH01012048
ODYU01006850
SOQ49066.1
JTDY01002062
KOB72202.1
NWSH01000902
+ More
PCG73616.1 KQ461190 KPJ07209.1 KQ459590 KPI97337.1 AGBW02010462 OWR48466.1 RSAL01000080 RVE48626.1 GEZM01077596 JAV63256.1 KQ977642 KYN00985.1 GAKP01010215 JAC48737.1 KQ982585 KYQ54259.1 GDHF01018156 JAI34158.1 AJVK01001814 KQ976625 KYM78982.1 GBXI01000925 JAD13367.1 AE014297 BT031026 AAF54521.1 ABV82408.1 GL448770 EFN83868.1 CP012526 ALC47053.1 KQ981979 KYN31516.1 GFXV01004690 MBW16495.1 GGMR01007555 MBY20174.1 KQ980295 KYN16858.1 KK853541 KDR06888.1 CH480815 EDW42741.1 CM000364 EDX13497.1 KQ435119 KZC14703.1 GL887491 EGI71028.1 ABLF02032259 LBMM01023662 KMQ82679.1 KZ288337 PBC27844.1 KQ414755 KOC61768.1 AK342037 BAH72224.1 ADTU01018578 GACK01007070 JAA57964.1 NEVH01013201 PNF29980.1 GEDV01007089 JAP81468.1 APGK01050879 APGK01050880 KB741178 ENN73168.1 OUUW01000013 SPP87694.1 BT128308 AEE63266.1 GFPF01003793 MAA14939.1 QOIP01000012 RLU16184.1 GGFL01000966 MBW65144.1 KK121581 KFM80778.1 KB632256 ERL90665.1 ACPB03013617 JRES01000752 KNC28754.1 GL761846 EFZ22680.1 KK854085 PTY10942.1 UYRR01031192 VDK47410.1 AXCM01010567 JXUM01042730 KQ561335 KXJ78931.1 GANO01000006 JAB59865.1 ATLV01023080 KE525340 KFB48425.1 APCN01004670 LN857024 CTP81947.1 JXUM01028011 KQ560807 KXJ80811.1 ADBV01000092 UYWW01003005 EJW88558.1 VDM12369.1 CH954181 EDV49640.1 AAAB01008948 EAA10420.4 AXCN02000973 JH429910 GFDL01003035 JAV32010.1 PYGN01000310 PSN48763.1 JH430221 UYRT01005362 VDK38550.1 JYDQ01000001 KRY23850.1 CH478041 EAT34237.1
PCG73616.1 KQ461190 KPJ07209.1 KQ459590 KPI97337.1 AGBW02010462 OWR48466.1 RSAL01000080 RVE48626.1 GEZM01077596 JAV63256.1 KQ977642 KYN00985.1 GAKP01010215 JAC48737.1 KQ982585 KYQ54259.1 GDHF01018156 JAI34158.1 AJVK01001814 KQ976625 KYM78982.1 GBXI01000925 JAD13367.1 AE014297 BT031026 AAF54521.1 ABV82408.1 GL448770 EFN83868.1 CP012526 ALC47053.1 KQ981979 KYN31516.1 GFXV01004690 MBW16495.1 GGMR01007555 MBY20174.1 KQ980295 KYN16858.1 KK853541 KDR06888.1 CH480815 EDW42741.1 CM000364 EDX13497.1 KQ435119 KZC14703.1 GL887491 EGI71028.1 ABLF02032259 LBMM01023662 KMQ82679.1 KZ288337 PBC27844.1 KQ414755 KOC61768.1 AK342037 BAH72224.1 ADTU01018578 GACK01007070 JAA57964.1 NEVH01013201 PNF29980.1 GEDV01007089 JAP81468.1 APGK01050879 APGK01050880 KB741178 ENN73168.1 OUUW01000013 SPP87694.1 BT128308 AEE63266.1 GFPF01003793 MAA14939.1 QOIP01000012 RLU16184.1 GGFL01000966 MBW65144.1 KK121581 KFM80778.1 KB632256 ERL90665.1 ACPB03013617 JRES01000752 KNC28754.1 GL761846 EFZ22680.1 KK854085 PTY10942.1 UYRR01031192 VDK47410.1 AXCM01010567 JXUM01042730 KQ561335 KXJ78931.1 GANO01000006 JAB59865.1 ATLV01023080 KE525340 KFB48425.1 APCN01004670 LN857024 CTP81947.1 JXUM01028011 KQ560807 KXJ80811.1 ADBV01000092 UYWW01003005 EJW88558.1 VDM12369.1 CH954181 EDV49640.1 AAAB01008948 EAA10420.4 AXCN02000973 JH429910 GFDL01003035 JAV32010.1 PYGN01000310 PSN48763.1 JH430221 UYRT01005362 VDK38550.1 JYDQ01000001 KRY23850.1 CH478041 EAT34237.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000283053 UP000075885 UP000075903 UP000078542 UP000075809 UP000092462 UP000095301 UP000091820 UP000078540 UP000000803 UP000008237 UP000092553 UP000078541 UP000078492 UP000027135 UP000001292 UP000000304 UP000076502 UP000007755 UP000007819 UP000095300 UP000036403 UP000242457 UP000053825 UP000005205 UP000235965 UP000019118 UP000268350 UP000279307 UP000075880 UP000005203 UP000054359 UP000030742 UP000075881 UP000015103 UP000037069 UP000036680 UP000075883 UP000069940 UP000249989 UP000030765 UP000075840 UP000006672 UP000004810 UP000093561 UP000270924 UP000008711 UP000075902 UP000007062 UP000076407 UP000075886 UP000245037 UP000050760 UP000054783 UP000008820
UP000283053 UP000075885 UP000075903 UP000078542 UP000075809 UP000092462 UP000095301 UP000091820 UP000078540 UP000000803 UP000008237 UP000092553 UP000078541 UP000078492 UP000027135 UP000001292 UP000000304 UP000076502 UP000007755 UP000007819 UP000095300 UP000036403 UP000242457 UP000053825 UP000005205 UP000235965 UP000019118 UP000268350 UP000279307 UP000075880 UP000005203 UP000054359 UP000030742 UP000075881 UP000015103 UP000037069 UP000036680 UP000075883 UP000069940 UP000249989 UP000030765 UP000075840 UP000006672 UP000004810 UP000093561 UP000270924 UP000008711 UP000075902 UP000007062 UP000076407 UP000075886 UP000245037 UP000050760 UP000054783 UP000008820
ProteinModelPortal
H9JQJ2
A0A2H1W7Y4
A0A0L7L9R1
A0A2A4JPD2
A0A194QPD1
A0A194PVI6
+ More
A0A212F3Z9 A0A3S2NZP6 A0A182PGA5 A0A1Y1KV23 A0A182V7P1 A0A195CJS7 A0A034W365 A0A151X1H8 A0A0K8V5X7 A0A1B0EYK1 A0A1I8N8N7 A0A1A9W2B1 A0A195B3Y5 A0A0A1XQZ4 Q9VH00 E2BK71 A0A0M3QY48 A0A195ETN0 A0A2H8TQS4 A0A2S2NSP7 A0A195DVF0 A0A067QGG2 B4HIU4 B4QV91 A0A154PS57 F4W432 J9K0A9 A0A1I8PJ24 A0A0J7JX82 A0A2A3E8Q5 A0A0L7QSY9 C4WWF4 A0A158NK02 L7M4D9 A0A2J7QN15 A0A131YSB0 N6T678 A0A3B0K066 J3JYI0 A0A224YGQ6 A0A3L8D7C5 A0A182IVF9 A0A088AAD8 A0A2M4CII5 A0A087UTT9 U4UKY2 A0A182JPV3 T1HPZ0 A0A0L0CBB0 E9I9T6 A0A2R7VTT8 A0A0M3JXB8 A0A182LZ25 A0A182G4Y7 U5EZW6 A0A084WDY1 A0A182HQN6 A0A0K0JMN7 A0A182H754 J9F255 B3P1P1 A0A182U7A6 Q7Q7W7 A0A182XJI1 A0A182QJ85 T1IHK6 A0A1Q3FWN0 A0A2P8YWY8 T1IJ04 A0A183D3C9 A0A0V1AH27 Q16IZ6
A0A212F3Z9 A0A3S2NZP6 A0A182PGA5 A0A1Y1KV23 A0A182V7P1 A0A195CJS7 A0A034W365 A0A151X1H8 A0A0K8V5X7 A0A1B0EYK1 A0A1I8N8N7 A0A1A9W2B1 A0A195B3Y5 A0A0A1XQZ4 Q9VH00 E2BK71 A0A0M3QY48 A0A195ETN0 A0A2H8TQS4 A0A2S2NSP7 A0A195DVF0 A0A067QGG2 B4HIU4 B4QV91 A0A154PS57 F4W432 J9K0A9 A0A1I8PJ24 A0A0J7JX82 A0A2A3E8Q5 A0A0L7QSY9 C4WWF4 A0A158NK02 L7M4D9 A0A2J7QN15 A0A131YSB0 N6T678 A0A3B0K066 J3JYI0 A0A224YGQ6 A0A3L8D7C5 A0A182IVF9 A0A088AAD8 A0A2M4CII5 A0A087UTT9 U4UKY2 A0A182JPV3 T1HPZ0 A0A0L0CBB0 E9I9T6 A0A2R7VTT8 A0A0M3JXB8 A0A182LZ25 A0A182G4Y7 U5EZW6 A0A084WDY1 A0A182HQN6 A0A0K0JMN7 A0A182H754 J9F255 B3P1P1 A0A182U7A6 Q7Q7W7 A0A182XJI1 A0A182QJ85 T1IHK6 A0A1Q3FWN0 A0A2P8YWY8 T1IJ04 A0A183D3C9 A0A0V1AH27 Q16IZ6
Ontologies
GO
PANTHER
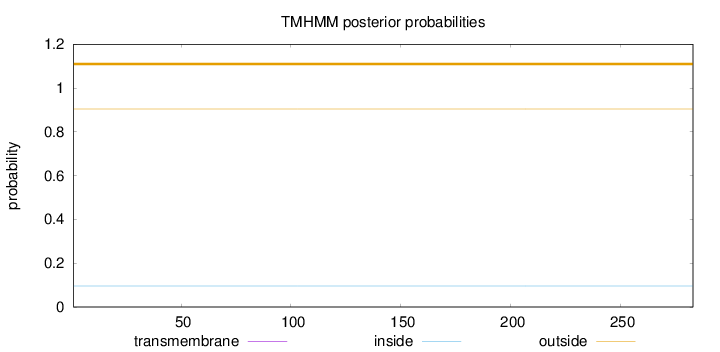
Topology
Length:
283
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09552
outside
1 - 283
Population Genetic Test Statistics
Pi
330.808105
Theta
218.56201
Tajima's D
1.581013
CLR
0
CSRT
0.8011599420029
Interpretation
Uncertain
