Pre Gene Modal
BGIBMGA011675
Annotation
Nuclear_pore_complex_protein_Nup160-like_[Papilio_xuthus]
Full name
Nuclear pore complex protein Nup160 homolog
Alternative Name
Nuclear pore protein 160
Location in the cell
PlasmaMembrane Reliability : 2.065
Sequence
CDS
ATGGAATTTCTTGAAGCTATGCCGGTCACGTTCAAGGAGATTGCTCCAAATCATAATATTCCAGAGAAATGGAAAGAAATTGTACTTAACACAGGTGGTACTCACAGTACTTTACAAGACATCAAATTACCGCAAAAAGCCGGCGGTTACTGGTATAAGGATTCAACGAAGTTCTACACCAGGAATCGCTTTATTTACTGGCAAACTACATCAGATGCAATTGAGATATCGGAAGCTTCACTAGATTTAAATCTCTCTTCTGCAAACTTGAGATGTAAAGTAGCTCCAGGAACTCCTCCACTGAGTAATATTACTGTTTATGAACGCAGTAGTACAAACAATGTTGTTATATTAGCTGCTACAGTTTCGTCAGTACATAGACTAGTTTTTCCACATCCTTCAATATTAGATAAAAAGTGTACATATGGATCAATGAGCAGTACATCACCTTCTATATGTCATGATACATCTTCTATAAACAACCCTAATAATTATTTTATTTTAAATCAGTATTCTAATGCCACCACTGGAGTAGCCCATACATGTTCTTCACTGCTTCGTGAAAATGGTGAAGCTATATTTGCACTTGCATTTGGACATGAAGGTGAGGGCAGTTTGTTACTTATAAAACTGCCAGTAACTGGAGCAGCAGTCACTACTGTATTAAAGAGAGAATCAACTGTACCCAGTTTCTTATCAGGAATTACAGGAGCTCTCAGAGGCAAGACTACTGATGGAGTTGAAACTTATGGTGTGGTAATTACAAAAAATGTAATTGTGGCTGTGTGTGGTGATGCGTGTCTTAGAGCATGGCCCATTGATGATGGAGGTGCACCAATTTCTGTGTCTGCCCCACTTTCACAAACAAACGTGAGACCGAAGCCACCACCACATGGTCACATGCTACAGAAAACAATTGGATTGGATGGCAGTGTCATTTTAGTTGCATATTTATCTTTTCCAAATGAATGTGAATTTGTTGTGATGAAGATGCATGATCCCGTAGGGGGATTTATAAAGTTTTCACATATCTGCCGAATATTTGGTCCTCAGTTGGATCTTTTAGATTATAAAATTGGTTATGGAGACAGCAATCATGTCATTTGGGCACTGTGGAGTCAACCCGATGGCGATACCATGATGACATGTAGTTCCCTGGGCGCCGAAGTGTCCTGGCAGGCAGTAGCGGGCCGCGAGCCTCCCCCGCCCTTGCCGCAACTATCAAATCACAACCAGTACCGCGATAGACTCCTTGCGCCGGGCCTATTTCCACATTCGGTCATCAGAAAAGCTCTGGTGATCTATGCGCGCACGTGGTGCGGCGGGGGCGCGGGGGGCGCGGCGGAGGAGGAGCTGGGCGAGGCGGCGGTGTCCGCCGTGCTGGCCCGCCTCCGCCTGCATGCGGCGCGCACCGCCTCCACAGATCAAACACATCTTATGCACAAATGCTGGTCAGATTTGTACAGCTGGTGCATGCAGTATATGGAAGGGTTACAGAAACCTCTTGGATTGATGGTGTCAGAAAATGGCACGGACATAGAGAGCGGAGGGTGGTGTGCAGTTGTGCGCCGCGCGGGGGTGTCTCTAGTCTGTGAACTCGAACCGCTAGAACGCATGATGTTGTCACCTGAAGATACACTGGAATCTGGAACTGGCGTGTGGCTGGGCGCGCTGTCGGCGGCGGCGCGGCGCGTGGTGGCGGCGGGCGCGTGCTGGGAGCGGGCCGCCTCGCCGAGCCGCGCGCACCAGCTCGAGCGACAACTGTTCGCAGCCGCCGCGCCGCAACACCGCCTGCTTCCTAGACTGCTGCATCTGCTGCACTCGCCGCCTATCTTAACACCCCAGCAAATCAATGAACTTAGTGCAATTTTGGAGCCGATCAGAGATCTCCAAAGTGCCGTACTGGAACTGAACGATGCCCTCAGATTGGATGTTCCAGACATCGAAATGTCTAGAAGTGAAGATGATGAAGACGGTGAATATGAAAGTTTATTTGCCAGCGATCTCGGAGTAGCCGTGGTCACCGAGGCCATTAGGCAAATGGCTGAAATGAGATGCCGCGTGGTGCGAGGCGCGCTGGCGGCGCTGGGCGTGTGTCGCGGGGCGGGCGCGGTTCCGGGCGCCGGACACTGCGCCGTGCACTGGCAGGCCTACCGCGCGCTTCTATGGCTGCACGGCGCGCCGCCACATGCCGCCACGAATTCGAATTCGTCGGAAGCGTTCCGGTTGAAGCTGAGTGCGCTAGGCGGCGCGGCAGTGAGTGCGGGCGCGGGCGGCGCGGGCGTGGTGCGGGTATACGCGCGGGGGCCGGGGGGCGCCCGGGCGCGCGCGCACCTGGCCGCCGCACGGACGCACACCGCCTGGCAACACGCGCTGCCGCACTTCCTCTACCATCTCGCGCATCAACTTTGGGCAGTTAGCGGCGGCTTCGATTTTGGTTGGTGGTTAGCAAGAATCAACCAGCCACGACTCGTGCAGACCTACGTCAATATGCTGGAGCCCTGGTGTGAATGGAATGCGTGCTCGCGTCAGTTTATATTGGGGTTGGCCCTGCTAGACCTGGAAGACACGGAGGCGGCGTACACGGCGTTCTGCAAGGCAGCCAAAGGTGTTAGCACGGAGCCTTTCCTGCGACACCTCGTGGCCGAGCCGGACGCGCACCTCACTCAGCACCGAGCCCTCGTGCTGTACTACATGAAGGTTATAAAATTGTTCGAGATTCACGACGAAGGCGCTTGCGTTGTGCGTCTCGCGGAAACCGCCATTAGCATCGCTGACAAAGATGATCCCAATTTGTCGATGTTCCAGTGGGTGGTGTTCAAATGGCACCTGTCGGGCGGGCGCACGTCCCGCGCGCTGACCGCCGCCGCCGCCAACCCGGCGCCGCACGCCCGCGCCGCCGCTGCGGCCGCCCTGCTCACTACGCTCGGTATATATACAATCTACCAATAG
Protein
MEFLEAMPVTFKEIAPNHNIPEKWKEIVLNTGGTHSTLQDIKLPQKAGGYWYKDSTKFYTRNRFIYWQTTSDAIEISEASLDLNLSSANLRCKVAPGTPPLSNITVYERSSTNNVVILAATVSSVHRLVFPHPSILDKKCTYGSMSSTSPSICHDTSSINNPNNYFILNQYSNATTGVAHTCSSLLRENGEAIFALAFGHEGEGSLLLIKLPVTGAAVTTVLKRESTVPSFLSGITGALRGKTTDGVETYGVVITKNVIVAVCGDACLRAWPIDDGGAPISVSAPLSQTNVRPKPPPHGHMLQKTIGLDGSVILVAYLSFPNECEFVVMKMHDPVGGFIKFSHICRIFGPQLDLLDYKIGYGDSNHVIWALWSQPDGDTMMTCSSLGAEVSWQAVAGREPPPPLPQLSNHNQYRDRLLAPGLFPHSVIRKALVIYARTWCGGGAGGAAEEELGEAAVSAVLARLRLHAARTASTDQTHLMHKCWSDLYSWCMQYMEGLQKPLGLMVSENGTDIESGGWCAVVRRAGVSLVCELEPLERMMLSPEDTLESGTGVWLGALSAAARRVVAAGACWERAASPSRAHQLERQLFAAAAPQHRLLPRLLHLLHSPPILTPQQINELSAILEPIRDLQSAVLELNDALRLDVPDIEMSRSEDDEDGEYESLFASDLGVAVVTEAIRQMAEMRCRVVRGALAALGVCRGAGAVPGAGHCAVHWQAYRALLWLHGAPPHAATNSNSSEAFRLKLSALGGAAVSAGAGGAGVVRVYARGPGGARARAHLAAARTHTAWQHALPHFLYHLAHQLWAVSGGFDFGWWLARINQPRLVQTYVNMLEPWCEWNACSRQFILGLALLDLEDTEAAYTAFCKAAKGVSTEPFLRHLVAEPDAHLTQHRALVLYYMKVIKLFEIHDEGACVVRLAETAISIADKDDPNLSMFQWVVFKWHLSGGRTSRALTAAAANPAPHARAAAAAALLTTLGIYTIYQ
Summary
Description
Involved in poly(A)+ RNA transport (By similarity). Required for nuclear import of Mad (PubMed:20547758). May play a role in double strand break DNA repair (PubMed:26502056).
Subunit
Interacts with Nup98, members of the Nup107 subcomplex in the nuclear pore.
Keywords
Complete proteome
DNA damage
mRNA transport
Nuclear pore complex
Nucleus
Polymorphism
Protein transport
Reference proteome
Translocation
Transport
Feature
chain Nuclear pore complex protein Nup160 homolog
Uniprot
S4PE43
A0A194PX43
A0A194QNW1
A0A2H1W1Q6
A0A212F427
A0A067RSR0
+ More
A0A2J7PQA4 A0A2J7PQB2 A0A0C9R7L3 A0A0C9QTL0 A0A0N0BCJ2 A0A087ZSU1 A0A2J7PQB1 A0A2P8Z5R9 E2B6Z5 A0A2A3E3A2 A0A1B0DPT4 D6X0C2 A0A1B0CGF9 A0A0L7RA15 A0A1I8PEF6 A0A1I8M5I5 A0A1A9W7W3 A0A1A9UV70 A0A158NN65 A0A195BB07 A0A1B0G1N8 A0A1A9ZLZ1 A0A195C422 A0A1B0B1N7 A0A1A9XHL8 A0A336LZC2 A0A336M2C2 A0A232FB08 A0A151X2K7 A0A195FAF0 A0A0H3XWL9 A0A0H3XSX0 A0A0H3XX57 A0A0H3XWM4 A0A0H3XX51 A0A0H3XTX4 A0A0H3XWE5 A0A0H3XWE1 C0KG82 A0A0H3XWF0 C0KG80 C0KG79 C0KG88 C0KG78 A0A0H3XTY4 B4P176 A0A0H3XWF5 C0KG87 C0KG86 C0KG77 C0KG81 C0KG84 X2J9F3 Q9VKJ3 B4HX22 A0A0H3XTX9 A0A1Y1M1R8 F4W8C9 A0A182GWQ8 A0A1S4F1D8 B0WAM0 A0A1Q3EZU2 B3N4K7
A0A2J7PQA4 A0A2J7PQB2 A0A0C9R7L3 A0A0C9QTL0 A0A0N0BCJ2 A0A087ZSU1 A0A2J7PQB1 A0A2P8Z5R9 E2B6Z5 A0A2A3E3A2 A0A1B0DPT4 D6X0C2 A0A1B0CGF9 A0A0L7RA15 A0A1I8PEF6 A0A1I8M5I5 A0A1A9W7W3 A0A1A9UV70 A0A158NN65 A0A195BB07 A0A1B0G1N8 A0A1A9ZLZ1 A0A195C422 A0A1B0B1N7 A0A1A9XHL8 A0A336LZC2 A0A336M2C2 A0A232FB08 A0A151X2K7 A0A195FAF0 A0A0H3XWL9 A0A0H3XSX0 A0A0H3XX57 A0A0H3XWM4 A0A0H3XX51 A0A0H3XTX4 A0A0H3XWE5 A0A0H3XWE1 C0KG82 A0A0H3XWF0 C0KG80 C0KG79 C0KG88 C0KG78 A0A0H3XTY4 B4P176 A0A0H3XWF5 C0KG87 C0KG86 C0KG77 C0KG81 C0KG84 X2J9F3 Q9VKJ3 B4HX22 A0A0H3XTX9 A0A1Y1M1R8 F4W8C9 A0A182GWQ8 A0A1S4F1D8 B0WAM0 A0A1Q3EZU2 B3N4K7
Pubmed
EMBL
GAIX01003336
JAA89224.1
KQ459590
KPI97334.1
KQ461190
KPJ07213.1
+ More
ODYU01005769 SOQ46967.1 AGBW02010462 OWR48463.1 KK852442 KDR23845.1 NEVH01022640 PNF18519.1 PNF18520.1 GBYB01004015 JAG73782.1 GBYB01004012 JAG73779.1 KQ435903 KOX69061.1 PNF18521.1 PYGN01000183 PSN51838.1 GL446071 EFN88537.1 KZ288446 PBC25619.1 AJVK01008241 AJVK01008242 KQ971372 EFA09598.2 AJWK01011081 KQ414619 KOC67705.1 ADTU01021054 KQ976537 KYM81417.1 CCAG010010949 KQ978344 KYM94923.1 JXJN01007216 UFQT01000316 SSX23160.1 SSX23159.1 NNAY01000491 OXU28021.1 KQ982579 KYQ54547.1 KQ981693 KYN37600.1 KR817629 AKM95868.1 KR817636 AKM95875.1 KR817635 AKM95874.1 KR817634 AKM95873.1 KR817630 KR817631 AKM95869.1 AKM95870.1 KR817628 AKM95867.1 KR817627 AKM95866.1 KR817621 KR817622 KR817623 KR817624 KR817625 KR817626 AKM95860.1 AKM95861.1 AKM95862.1 AKM95863.1 AKM95864.1 AKM95865.1 FJ600395 ACM79354.1 KR817632 AKM95871.1 FJ600393 FJ600396 CM002910 ACM79352.1 ACM79355.1 KMY89705.1 FJ600392 ACM79351.1 FJ600401 ACM79360.1 FJ600391 ACM79350.1 KR817638 AKM95877.1 CM000157 EDW88051.1 KR817637 AKM95876.1 FJ600400 ACM79359.1 FJ600399 ACM79358.1 FJ600390 ACM79349.1 FJ600394 ACM79353.1 FJ600397 FJ600398 ACM79356.1 ACM79357.1 AE014134 AHN54346.1 FJ600378 FJ600379 FJ600380 FJ600381 FJ600382 FJ600383 FJ600384 FJ600385 FJ600386 FJ600387 FJ600388 FJ600389 BT015261 CH480818 EDW52567.1 KR817633 AKM95872.1 GEZM01042736 GEZM01042735 JAV79669.1 GL887908 EGI69420.1 JXUM01093892 JXUM01093893 JXUM01093894 KQ564087 KXJ72815.1 DS231873 EDS41574.1 GFDL01014222 JAV20823.1 CH954177 EDV58919.1
ODYU01005769 SOQ46967.1 AGBW02010462 OWR48463.1 KK852442 KDR23845.1 NEVH01022640 PNF18519.1 PNF18520.1 GBYB01004015 JAG73782.1 GBYB01004012 JAG73779.1 KQ435903 KOX69061.1 PNF18521.1 PYGN01000183 PSN51838.1 GL446071 EFN88537.1 KZ288446 PBC25619.1 AJVK01008241 AJVK01008242 KQ971372 EFA09598.2 AJWK01011081 KQ414619 KOC67705.1 ADTU01021054 KQ976537 KYM81417.1 CCAG010010949 KQ978344 KYM94923.1 JXJN01007216 UFQT01000316 SSX23160.1 SSX23159.1 NNAY01000491 OXU28021.1 KQ982579 KYQ54547.1 KQ981693 KYN37600.1 KR817629 AKM95868.1 KR817636 AKM95875.1 KR817635 AKM95874.1 KR817634 AKM95873.1 KR817630 KR817631 AKM95869.1 AKM95870.1 KR817628 AKM95867.1 KR817627 AKM95866.1 KR817621 KR817622 KR817623 KR817624 KR817625 KR817626 AKM95860.1 AKM95861.1 AKM95862.1 AKM95863.1 AKM95864.1 AKM95865.1 FJ600395 ACM79354.1 KR817632 AKM95871.1 FJ600393 FJ600396 CM002910 ACM79352.1 ACM79355.1 KMY89705.1 FJ600392 ACM79351.1 FJ600401 ACM79360.1 FJ600391 ACM79350.1 KR817638 AKM95877.1 CM000157 EDW88051.1 KR817637 AKM95876.1 FJ600400 ACM79359.1 FJ600399 ACM79358.1 FJ600390 ACM79349.1 FJ600394 ACM79353.1 FJ600397 FJ600398 ACM79356.1 ACM79357.1 AE014134 AHN54346.1 FJ600378 FJ600379 FJ600380 FJ600381 FJ600382 FJ600383 FJ600384 FJ600385 FJ600386 FJ600387 FJ600388 FJ600389 BT015261 CH480818 EDW52567.1 KR817633 AKM95872.1 GEZM01042736 GEZM01042735 JAV79669.1 GL887908 EGI69420.1 JXUM01093892 JXUM01093893 JXUM01093894 KQ564087 KXJ72815.1 DS231873 EDS41574.1 GFDL01014222 JAV20823.1 CH954177 EDV58919.1
Proteomes
UP000053268
UP000053240
UP000007151
UP000027135
UP000235965
UP000053105
+ More
UP000005203 UP000245037 UP000008237 UP000242457 UP000092462 UP000007266 UP000092461 UP000053825 UP000095300 UP000095301 UP000091820 UP000078200 UP000005205 UP000078540 UP000092444 UP000092445 UP000078542 UP000092460 UP000092443 UP000215335 UP000075809 UP000078541 UP000002282 UP000000803 UP000001292 UP000007755 UP000069940 UP000249989 UP000002320 UP000008711
UP000005203 UP000245037 UP000008237 UP000242457 UP000092462 UP000007266 UP000092461 UP000053825 UP000095300 UP000095301 UP000091820 UP000078200 UP000005205 UP000078540 UP000092444 UP000092445 UP000078542 UP000092460 UP000092443 UP000215335 UP000075809 UP000078541 UP000002282 UP000000803 UP000001292 UP000007755 UP000069940 UP000249989 UP000002320 UP000008711
SUPFAM
SSF50978
SSF50978
ProteinModelPortal
S4PE43
A0A194PX43
A0A194QNW1
A0A2H1W1Q6
A0A212F427
A0A067RSR0
+ More
A0A2J7PQA4 A0A2J7PQB2 A0A0C9R7L3 A0A0C9QTL0 A0A0N0BCJ2 A0A087ZSU1 A0A2J7PQB1 A0A2P8Z5R9 E2B6Z5 A0A2A3E3A2 A0A1B0DPT4 D6X0C2 A0A1B0CGF9 A0A0L7RA15 A0A1I8PEF6 A0A1I8M5I5 A0A1A9W7W3 A0A1A9UV70 A0A158NN65 A0A195BB07 A0A1B0G1N8 A0A1A9ZLZ1 A0A195C422 A0A1B0B1N7 A0A1A9XHL8 A0A336LZC2 A0A336M2C2 A0A232FB08 A0A151X2K7 A0A195FAF0 A0A0H3XWL9 A0A0H3XSX0 A0A0H3XX57 A0A0H3XWM4 A0A0H3XX51 A0A0H3XTX4 A0A0H3XWE5 A0A0H3XWE1 C0KG82 A0A0H3XWF0 C0KG80 C0KG79 C0KG88 C0KG78 A0A0H3XTY4 B4P176 A0A0H3XWF5 C0KG87 C0KG86 C0KG77 C0KG81 C0KG84 X2J9F3 Q9VKJ3 B4HX22 A0A0H3XTX9 A0A1Y1M1R8 F4W8C9 A0A182GWQ8 A0A1S4F1D8 B0WAM0 A0A1Q3EZU2 B3N4K7
A0A2J7PQA4 A0A2J7PQB2 A0A0C9R7L3 A0A0C9QTL0 A0A0N0BCJ2 A0A087ZSU1 A0A2J7PQB1 A0A2P8Z5R9 E2B6Z5 A0A2A3E3A2 A0A1B0DPT4 D6X0C2 A0A1B0CGF9 A0A0L7RA15 A0A1I8PEF6 A0A1I8M5I5 A0A1A9W7W3 A0A1A9UV70 A0A158NN65 A0A195BB07 A0A1B0G1N8 A0A1A9ZLZ1 A0A195C422 A0A1B0B1N7 A0A1A9XHL8 A0A336LZC2 A0A336M2C2 A0A232FB08 A0A151X2K7 A0A195FAF0 A0A0H3XWL9 A0A0H3XSX0 A0A0H3XX57 A0A0H3XWM4 A0A0H3XX51 A0A0H3XTX4 A0A0H3XWE5 A0A0H3XWE1 C0KG82 A0A0H3XWF0 C0KG80 C0KG79 C0KG88 C0KG78 A0A0H3XTY4 B4P176 A0A0H3XWF5 C0KG87 C0KG86 C0KG77 C0KG81 C0KG84 X2J9F3 Q9VKJ3 B4HX22 A0A0H3XTX9 A0A1Y1M1R8 F4W8C9 A0A182GWQ8 A0A1S4F1D8 B0WAM0 A0A1Q3EZU2 B3N4K7
PDB
5A9Q
E-value=1.85902e-28,
Score=317
Ontologies
PANTHER
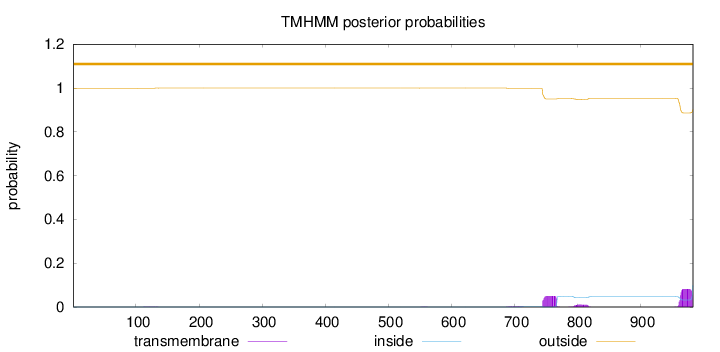
Topology
Subcellular location
Length:
983
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.99294
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00054
outside
1 - 983
Population Genetic Test Statistics
Pi
234.044576
Theta
182.116169
Tajima's D
1.009952
CLR
0.009629
CSRT
0.662916854157292
Interpretation
Uncertain
