Gene
KWMTBOMO06595 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011674
Annotation
puromycin-sensitive_aminopeptidase-like_[Bombyx_mori]
Full name
Aminopeptidase
Location in the cell
Cytoplasmic Reliability : 2.356
Sequence
CDS
ATGCCAGAGAACAAGCCGTTTCAACGACTTCCTAATAATGTGATTCCTAAGCATTATGCATTGGAATTGATTCCAAATCTGGAAAAATTTACTTTCAAGGGAAAAACTGCGGTGAAAGTATCGATTGTTAATCCAACGAATGTGATAGTATTGAATAGCTTAGACTTGGATTTGAAAAATGTGAAACTTCAGTATAACGATGGCTCTAATTCAGCGATAATTCCATCGTCAGTAGAATTGAGTACAACTGATGAAACAGCAAGTATTTATTTCTCAGAATCTTTACTAGAAGGCGAAGCGACGCTTTATTCTGAGTTTACAGGTGAAATAAATGACAAAATGAAAGGCTTGTACCGCAGTAAATACATTGCTCCCAATGGAGAAGAACGATATGCGGCTGTCACCCAATTTGAAGCGACAGATGCACGACGATGTTTCCCTTGCTGGGATGAACCTGCAATTAAAGCTACTTTTGATATAACCTTACAAGTTCCGGCTGATCGTGTTGCTTTGTCTAATATGCCTGTAAAACAAGAAAAGATTGCAGACAATACAAGGATCATACAATTTGACACAACACCAATAATGTCAACATATTTAGTGGCTGTTGTTGTTGGTGAATATGATTATGTGGAAAAGAAATCTAATGATGGAATTCTAGTAAGGGTTTATACTCCTGTAGGCAAAAGTAAACAGGGGTTGTTTGCACTTGAAGTGGCTGCACGAGTTTTGCCTTATTATAAAGATTACTTTGATATTGCATACCCCTTGCCCAAAATAGACCTGATTGCTATAGCAGACTTTTCAGCAGGAGCTATGGAGAATTGGGGATTAGTAACATATAGGGAGACCTGTCTCTTGGTAGATGAAGAACATACTTCTGCTGTTAGAAGACAATGGATAGCTTTAGTTGTAGGACATGAACTGGCACATCAATGGTTTGGCAATCTTGTAACAATGGAATGGTGGACACATCTCTGGCTGAATGAAGGATATGCATCTTTTGTGGAATTTCTTTGTGTAAATCATCTTTTCCCAGAATATGATATTTGGACTCAGTTTGTGACTGAAAACTACATTAGAGCATTGGAATTGGATTGCTTAAAAAATTCACACCCAATTGAGGTTCCTGTTGGCCATCCATCAGAAATTGATGAAATATTTGATGACATATCGTACAATAAGGGGGCATCTGTAATACGTATGTTACATAGATATATTGGCGATGAAGATTTTCGTAAGGGCATGAATATCTACCTAACTCGTCATCAGTACAAGAACACTTTCACTGAAGATTTGTGGGCAGCACTGGAAGAGGCGTCTAAAAAGCCTGTCGGAGCTGTTATGTCTACATGGACCAAACAGATGGGTTTCCCGATGGTTGAGGTTCAGTCAGAACAGCGGGGCTCCAATCGAGTATTAACATTAACTCAACGCAAATTCTGTGCGGATGGTAGTCAAGCAGATGACACTCTTTGGATGGTTCCAATATCCATATCAACGCAAGAACAACCTTCAAAGGTAGCTTTGTCTATGGTTTTAGAAAAGAGAACACAGGAAGTTGTATTGAAAAATGTAGCTCAAGACTCTTGGATTAAATTGAATCCAGGAACTGTGGGGTACTATCGCACACGGTACCCGGCAGAGCTGTTGGAGCAGCTGGTGCCGGCCATACGGGATGGCTCTCTGCCGCCGCTCGACCGCCTCGGACTACTGGACGACTGCTTCGCACTCGTGCAAGCGGGACACACGCACACTGCCGATTCTTTGAAACTTATGGAAGCATTCAGCAATGAAACAAATTTTACTGTGTGGTCGACCATCGCCAATTGTATGTCTAAGCTAAGTGCACTATTTTCTCAAACTGCTCTCGACAAACCCTTGAAAAATTACGGACGAAAGCTATTTTCAAACATAACCAAGAAATTGGGATGGGACGCGGAAGAGAAAGAAAGTCACCTCGACACTCTGCTCAGAAGCTTGGTGCTCAATAAGATGATTAGCTTTGAAGACCCTGACACCATTAAAGAAGCGAAAATTCGGTTTGAGAAGCACATTTCAGGCGAGCGGCCGCTGGCGGCGGACCTGCGCTCGGCGTGCTACCGCGCGGAGCTGGGCGGCGCCGACGAGCGCGTGTTCGAGCGGTTCCTGCAACTGTACCGCGCCGCCGACCTGCACGAGGAGAAGGACCGCGTGTCGCGGGCCCTGGGCGCCGTGCGGGACCCCGCGCTGCTCAGGCGCGTGCTCGACTTTGCCATCTCGGATGAAGTAAGATCGCAGGACACAGTTTTCGTTATCGTGTCAGTTGCAGTAAGCAGAAACGGTCGTGACCTGGCATGGCAGTTTTTCAAAGATCACTTCCAGGAATTCATAGAAAGATATCAGGGTGGATTCTTGTTGGCTCGGTTGGTGAAATCGACAACTGAAAACTTCGCGTCGGAGGCAGCAGCACAAGAGATTGAAGAGTTCTTCAGCACGCACGAGTCTCCCGGGGCGGAGCGCTCTGTTCAGCAGGCATTAGAGAGCGTTCGACTCAACGCGGCCTGGCTGCGCCGCGACCTCGCCTCCACCGCCACCTACCTGCACCCCTACCACTGA
Protein
MPENKPFQRLPNNVIPKHYALELIPNLEKFTFKGKTAVKVSIVNPTNVIVLNSLDLDLKNVKLQYNDGSNSAIIPSSVELSTTDETASIYFSESLLEGEATLYSEFTGEINDKMKGLYRSKYIAPNGEERYAAVTQFEATDARRCFPCWDEPAIKATFDITLQVPADRVALSNMPVKQEKIADNTRIIQFDTTPIMSTYLVAVVVGEYDYVEKKSNDGILVRVYTPVGKSKQGLFALEVAARVLPYYKDYFDIAYPLPKIDLIAIADFSAGAMENWGLVTYRETCLLVDEEHTSAVRRQWIALVVGHELAHQWFGNLVTMEWWTHLWLNEGYASFVEFLCVNHLFPEYDIWTQFVTENYIRALELDCLKNSHPIEVPVGHPSEIDEIFDDISYNKGASVIRMLHRYIGDEDFRKGMNIYLTRHQYKNTFTEDLWAALEEASKKPVGAVMSTWTKQMGFPMVEVQSEQRGSNRVLTLTQRKFCADGSQADDTLWMVPISISTQEQPSKVALSMVLEKRTQEVVLKNVAQDSWIKLNPGTVGYYRTRYPAELLEQLVPAIRDGSLPPLDRLGLLDDCFALVQAGHTHTADSLKLMEAFSNETNFTVWSTIANCMSKLSALFSQTALDKPLKNYGRKLFSNITKKLGWDAEEKESHLDTLLRSLVLNKMISFEDPDTIKEAKIRFEKHISGERPLAADLRSACYRAELGGADERVFERFLQLYRAADLHEEKDRVSRALGAVRDPALLRRVLDFAISDEVRSQDTVFVIVSVAVSRNGRDLAWQFFKDHFQEFIERYQGGFLLARLVKSTTENFASEAAAQEIEEFFSTHESPGAERSVQQALESVRLNAAWLRRDLASTATYLHPYH
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M1 family.
Uniprot
I3VR81
H9JQ68
A0A212F406
A0A2W1BLC5
A0A2H1W1L9
S4PRY9
+ More
A0A067R998 A0A1W4WKI4 A0A2J7PQP5 E0VR15 A0A0K8TTN7 A0A0R1E0N1 B4QMF5 A0A0R1DUK5 A0A0R1DZS0 B4PD97 A0A0R1DU86 A0A0T6B7U7 A0A1L8E2T5 A0A0J9UC25 A0A1W4VU03 A0A1Y1LCB5 A0A1L8E2H5 A0A0J9RN50 A0A1W4W6K8 A0A1W4VU07 B4HVU3 A0A0J9RLM8 Q8IRH1 A0A1W4W577 E8NH92 Q8IRH0 A0A0Q5UCX3 B3NBB7 Q9W0E4 A0A0Q5U1R8 C8VUZ1 A0A0Q9WHK8 B4LCX5 A0A182QNL7 A0A0M3QW75 Q5TRG5 A0A182V0Y7 A0A3B0K9Q6 A0A3B0KIE7 W8ANG5 Q29FE8 B4H1F6 Q9GPG3 A0A0P4WFE2 A0A0R3P2G6 W8AJQ4 A0A0A1X6F9 A0A084VPY2 A0A0A1WT76 B4IXJ3 A0A0P9C240 A0A0P8YFL8 A0A0P8XT10 B3M8F9 A0A182RIS4 B4L8V4 A0A1I8PNJ7 B4MKU7 A0A1I8PNJ8 A0A034VU83 A0A034VSE7 A0A0L0CND7 A0A087ZTK0 A0A1I8MF11 A0A0K8VNJ6 T1PIH7 A0A1B6FHQ9 A0A1A9UGB1 A0A195CUU7 A0A1B0G3B9 K7JA89 A0A0C9PNN1 A0A1B6E9J3 A0A1B0B6Z2 A0A1A9YIB9 A0A2M4CK94 A0A2M4CKA0 E2BM61 A0A2M4CKS9 A0A1A9WA91 A0A0A9YBG9 A0A2M3Z0D2 A0A2M4A3P0 A0A2M4BCC3 A0A2M4BBR0 A0A2M4A3V7 A0A2M4BD12 E2A5M2 A0A182JLL4 A0A1D2N281 A0A158NA38 A0A0P4VXS7 A0A0J7L546 A0A195FRL0
A0A067R998 A0A1W4WKI4 A0A2J7PQP5 E0VR15 A0A0K8TTN7 A0A0R1E0N1 B4QMF5 A0A0R1DUK5 A0A0R1DZS0 B4PD97 A0A0R1DU86 A0A0T6B7U7 A0A1L8E2T5 A0A0J9UC25 A0A1W4VU03 A0A1Y1LCB5 A0A1L8E2H5 A0A0J9RN50 A0A1W4W6K8 A0A1W4VU07 B4HVU3 A0A0J9RLM8 Q8IRH1 A0A1W4W577 E8NH92 Q8IRH0 A0A0Q5UCX3 B3NBB7 Q9W0E4 A0A0Q5U1R8 C8VUZ1 A0A0Q9WHK8 B4LCX5 A0A182QNL7 A0A0M3QW75 Q5TRG5 A0A182V0Y7 A0A3B0K9Q6 A0A3B0KIE7 W8ANG5 Q29FE8 B4H1F6 Q9GPG3 A0A0P4WFE2 A0A0R3P2G6 W8AJQ4 A0A0A1X6F9 A0A084VPY2 A0A0A1WT76 B4IXJ3 A0A0P9C240 A0A0P8YFL8 A0A0P8XT10 B3M8F9 A0A182RIS4 B4L8V4 A0A1I8PNJ7 B4MKU7 A0A1I8PNJ8 A0A034VU83 A0A034VSE7 A0A0L0CND7 A0A087ZTK0 A0A1I8MF11 A0A0K8VNJ6 T1PIH7 A0A1B6FHQ9 A0A1A9UGB1 A0A195CUU7 A0A1B0G3B9 K7JA89 A0A0C9PNN1 A0A1B6E9J3 A0A1B0B6Z2 A0A1A9YIB9 A0A2M4CK94 A0A2M4CKA0 E2BM61 A0A2M4CKS9 A0A1A9WA91 A0A0A9YBG9 A0A2M3Z0D2 A0A2M4A3P0 A0A2M4BCC3 A0A2M4BBR0 A0A2M4A3V7 A0A2M4BD12 E2A5M2 A0A182JLL4 A0A1D2N281 A0A158NA38 A0A0P4VXS7 A0A0J7L546 A0A195FRL0
EC Number
3.4.11.-
Pubmed
19121390
22118469
28756777
23622113
24845553
20566863
+ More
26369729 17994087 17550304 22936249 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 12364791 14747013 17210077 24495485 15632085 11819115 23185243 25830018 24438588 25348373 26108605 25315136 20075255 20798317 25401762 27289101 21347285 27129103
26369729 17994087 17550304 22936249 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 12364791 14747013 17210077 24495485 15632085 11819115 23185243 25830018 24438588 25348373 26108605 25315136 20075255 20798317 25401762 27289101 21347285 27129103
EMBL
JQ061152
AFK85026.1
BABH01012063
AGBW02010462
OWR48461.1
KZ150049
+ More
PZC74444.1 ODYU01005769 SOQ46969.1 GAIX01000390 JAA92170.1 KK852851 KDR15061.1 NEVH01022635 PNF18664.1 DS235442 EEB15821.1 GDAI01000090 JAI17513.1 CM000159 KRK00743.1 CM000363 EDX08812.1 KRK00741.1 KRK00746.1 EDW92845.1 KRK00742.1 LJIG01009289 KRT83377.1 GFDF01001145 JAV12939.1 CM002912 KMY96835.1 GEZM01063210 JAV69196.1 GFDF01001144 JAV12940.1 KMY96839.1 CH480817 EDW50058.1 KMY96838.1 AE014296 BT133253 AAN11480.1 AFB71128.1 BT125968 ADX35947.1 BT099693 AAN11481.1 ACV53057.1 CH954178 KQS43073.1 EDV50170.1 AAF47504.1 KQS43071.1 BT099737 ACV53871.1 CH940647 KRF84040.1 EDW68836.2 AXCN02000881 CP012525 ALC43653.1 AAAB01008960 EAL39899.3 OUUW01000009 SPP84860.1 SPP84861.1 GAMC01020567 JAB85988.1 CH379067 EAL31307.3 CH479202 EDW30133.1 AF327435 AAG48733.1 GDRN01034234 JAI67458.1 KRT07386.1 GAMC01020568 JAB85987.1 GBXI01007570 JAD06722.1 ATLV01015079 KE525001 KFB40026.1 GBXI01012491 JAD01801.1 CH916366 EDV96430.1 CH902618 KPU77721.1 KPU77720.1 KPU77719.1 EDV38894.1 CH933816 EDW17129.2 CH963847 EDW72803.2 GAKP01013557 JAC45395.1 GAKP01013558 JAC45394.1 JRES01000254 KNC32934.1 GDHF01011858 JAI40456.1 KA647733 AFP62362.1 GECZ01020049 JAS49720.1 KQ977276 KYN04456.1 CCAG010013759 AAZX01015428 GBYB01002728 JAG72495.1 GEDC01015936 GEDC01002690 JAS21362.1 JAS34608.1 JXJN01009324 GGFL01001604 MBW65782.1 GGFL01001602 MBW65780.1 GL449158 EFN83187.1 GGFL01001603 MBW65781.1 GBHO01016724 JAG26880.1 GGFM01001190 MBW21941.1 GGFK01002115 MBW35436.1 GGFJ01001327 MBW50468.1 GGFJ01001319 MBW50460.1 GGFK01002114 MBW35435.1 GGFJ01001798 MBW50939.1 GL437023 EFN71265.1 LJIJ01000285 ODM99382.1 ADTU01009957 GDKW01001138 JAI55457.1 LBMM01000742 KMQ97594.1 KQ981305 KYN42922.1
PZC74444.1 ODYU01005769 SOQ46969.1 GAIX01000390 JAA92170.1 KK852851 KDR15061.1 NEVH01022635 PNF18664.1 DS235442 EEB15821.1 GDAI01000090 JAI17513.1 CM000159 KRK00743.1 CM000363 EDX08812.1 KRK00741.1 KRK00746.1 EDW92845.1 KRK00742.1 LJIG01009289 KRT83377.1 GFDF01001145 JAV12939.1 CM002912 KMY96835.1 GEZM01063210 JAV69196.1 GFDF01001144 JAV12940.1 KMY96839.1 CH480817 EDW50058.1 KMY96838.1 AE014296 BT133253 AAN11480.1 AFB71128.1 BT125968 ADX35947.1 BT099693 AAN11481.1 ACV53057.1 CH954178 KQS43073.1 EDV50170.1 AAF47504.1 KQS43071.1 BT099737 ACV53871.1 CH940647 KRF84040.1 EDW68836.2 AXCN02000881 CP012525 ALC43653.1 AAAB01008960 EAL39899.3 OUUW01000009 SPP84860.1 SPP84861.1 GAMC01020567 JAB85988.1 CH379067 EAL31307.3 CH479202 EDW30133.1 AF327435 AAG48733.1 GDRN01034234 JAI67458.1 KRT07386.1 GAMC01020568 JAB85987.1 GBXI01007570 JAD06722.1 ATLV01015079 KE525001 KFB40026.1 GBXI01012491 JAD01801.1 CH916366 EDV96430.1 CH902618 KPU77721.1 KPU77720.1 KPU77719.1 EDV38894.1 CH933816 EDW17129.2 CH963847 EDW72803.2 GAKP01013557 JAC45395.1 GAKP01013558 JAC45394.1 JRES01000254 KNC32934.1 GDHF01011858 JAI40456.1 KA647733 AFP62362.1 GECZ01020049 JAS49720.1 KQ977276 KYN04456.1 CCAG010013759 AAZX01015428 GBYB01002728 JAG72495.1 GEDC01015936 GEDC01002690 JAS21362.1 JAS34608.1 JXJN01009324 GGFL01001604 MBW65782.1 GGFL01001602 MBW65780.1 GL449158 EFN83187.1 GGFL01001603 MBW65781.1 GBHO01016724 JAG26880.1 GGFM01001190 MBW21941.1 GGFK01002115 MBW35436.1 GGFJ01001327 MBW50468.1 GGFJ01001319 MBW50460.1 GGFK01002114 MBW35435.1 GGFJ01001798 MBW50939.1 GL437023 EFN71265.1 LJIJ01000285 ODM99382.1 ADTU01009957 GDKW01001138 JAI55457.1 LBMM01000742 KMQ97594.1 KQ981305 KYN42922.1
Proteomes
UP000005204
UP000007151
UP000027135
UP000192223
UP000235965
UP000009046
+ More
UP000002282 UP000000304 UP000192221 UP000001292 UP000000803 UP000008711 UP000008792 UP000075886 UP000092553 UP000007062 UP000075903 UP000268350 UP000001819 UP000008744 UP000030765 UP000001070 UP000007801 UP000075900 UP000009192 UP000095300 UP000007798 UP000037069 UP000005203 UP000095301 UP000078200 UP000078542 UP000092444 UP000002358 UP000092460 UP000092443 UP000008237 UP000091820 UP000000311 UP000075880 UP000094527 UP000005205 UP000036403 UP000078541
UP000002282 UP000000304 UP000192221 UP000001292 UP000000803 UP000008711 UP000008792 UP000075886 UP000092553 UP000007062 UP000075903 UP000268350 UP000001819 UP000008744 UP000030765 UP000001070 UP000007801 UP000075900 UP000009192 UP000095300 UP000007798 UP000037069 UP000005203 UP000095301 UP000078200 UP000078542 UP000092444 UP000002358 UP000092460 UP000092443 UP000008237 UP000091820 UP000000311 UP000075880 UP000094527 UP000005205 UP000036403 UP000078541
Interpro
Gene 3D
CDD
ProteinModelPortal
I3VR81
H9JQ68
A0A212F406
A0A2W1BLC5
A0A2H1W1L9
S4PRY9
+ More
A0A067R998 A0A1W4WKI4 A0A2J7PQP5 E0VR15 A0A0K8TTN7 A0A0R1E0N1 B4QMF5 A0A0R1DUK5 A0A0R1DZS0 B4PD97 A0A0R1DU86 A0A0T6B7U7 A0A1L8E2T5 A0A0J9UC25 A0A1W4VU03 A0A1Y1LCB5 A0A1L8E2H5 A0A0J9RN50 A0A1W4W6K8 A0A1W4VU07 B4HVU3 A0A0J9RLM8 Q8IRH1 A0A1W4W577 E8NH92 Q8IRH0 A0A0Q5UCX3 B3NBB7 Q9W0E4 A0A0Q5U1R8 C8VUZ1 A0A0Q9WHK8 B4LCX5 A0A182QNL7 A0A0M3QW75 Q5TRG5 A0A182V0Y7 A0A3B0K9Q6 A0A3B0KIE7 W8ANG5 Q29FE8 B4H1F6 Q9GPG3 A0A0P4WFE2 A0A0R3P2G6 W8AJQ4 A0A0A1X6F9 A0A084VPY2 A0A0A1WT76 B4IXJ3 A0A0P9C240 A0A0P8YFL8 A0A0P8XT10 B3M8F9 A0A182RIS4 B4L8V4 A0A1I8PNJ7 B4MKU7 A0A1I8PNJ8 A0A034VU83 A0A034VSE7 A0A0L0CND7 A0A087ZTK0 A0A1I8MF11 A0A0K8VNJ6 T1PIH7 A0A1B6FHQ9 A0A1A9UGB1 A0A195CUU7 A0A1B0G3B9 K7JA89 A0A0C9PNN1 A0A1B6E9J3 A0A1B0B6Z2 A0A1A9YIB9 A0A2M4CK94 A0A2M4CKA0 E2BM61 A0A2M4CKS9 A0A1A9WA91 A0A0A9YBG9 A0A2M3Z0D2 A0A2M4A3P0 A0A2M4BCC3 A0A2M4BBR0 A0A2M4A3V7 A0A2M4BD12 E2A5M2 A0A182JLL4 A0A1D2N281 A0A158NA38 A0A0P4VXS7 A0A0J7L546 A0A195FRL0
A0A067R998 A0A1W4WKI4 A0A2J7PQP5 E0VR15 A0A0K8TTN7 A0A0R1E0N1 B4QMF5 A0A0R1DUK5 A0A0R1DZS0 B4PD97 A0A0R1DU86 A0A0T6B7U7 A0A1L8E2T5 A0A0J9UC25 A0A1W4VU03 A0A1Y1LCB5 A0A1L8E2H5 A0A0J9RN50 A0A1W4W6K8 A0A1W4VU07 B4HVU3 A0A0J9RLM8 Q8IRH1 A0A1W4W577 E8NH92 Q8IRH0 A0A0Q5UCX3 B3NBB7 Q9W0E4 A0A0Q5U1R8 C8VUZ1 A0A0Q9WHK8 B4LCX5 A0A182QNL7 A0A0M3QW75 Q5TRG5 A0A182V0Y7 A0A3B0K9Q6 A0A3B0KIE7 W8ANG5 Q29FE8 B4H1F6 Q9GPG3 A0A0P4WFE2 A0A0R3P2G6 W8AJQ4 A0A0A1X6F9 A0A084VPY2 A0A0A1WT76 B4IXJ3 A0A0P9C240 A0A0P8YFL8 A0A0P8XT10 B3M8F9 A0A182RIS4 B4L8V4 A0A1I8PNJ7 B4MKU7 A0A1I8PNJ8 A0A034VU83 A0A034VSE7 A0A0L0CND7 A0A087ZTK0 A0A1I8MF11 A0A0K8VNJ6 T1PIH7 A0A1B6FHQ9 A0A1A9UGB1 A0A195CUU7 A0A1B0G3B9 K7JA89 A0A0C9PNN1 A0A1B6E9J3 A0A1B0B6Z2 A0A1A9YIB9 A0A2M4CK94 A0A2M4CKA0 E2BM61 A0A2M4CKS9 A0A1A9WA91 A0A0A9YBG9 A0A2M3Z0D2 A0A2M4A3P0 A0A2M4BCC3 A0A2M4BBR0 A0A2M4A3V7 A0A2M4BD12 E2A5M2 A0A182JLL4 A0A1D2N281 A0A158NA38 A0A0P4VXS7 A0A0J7L546 A0A195FRL0
PDB
4KXD
E-value=6.38405e-132,
Score=1209
Ontologies
GO
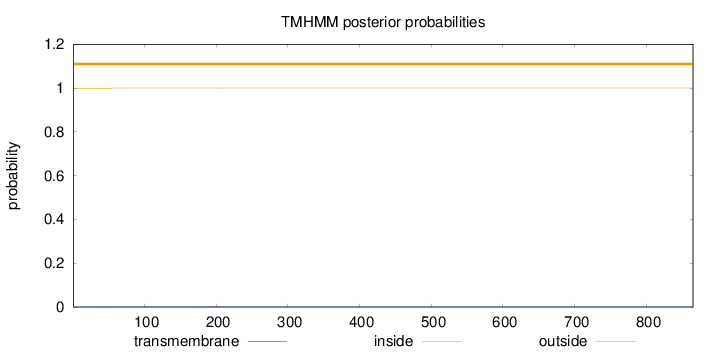
Topology
Length:
865
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01069
Exp number, first 60 AAs:
0.00201
Total prob of N-in:
0.00041
outside
1 - 865
Population Genetic Test Statistics
Pi
216.332509
Theta
159.640394
Tajima's D
1.264571
CLR
0
CSRT
0.731563421828909
Interpretation
Uncertain
