Gene
KWMTBOMO06592
Pre Gene Modal
BGIBMGA011673
Annotation
PREDICTED:_SNF-related_serine/threonine-protein_kinase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.174
Sequence
CDS
ATGGCAGTGAATAACAATGGCACTAGTGTAGCCAGTGGCACATATGACACAAAAATAGCGGGATTATATGACTTACTTGACACTCTTGGTTCAGGACATTTCGCTGTTGTAAAGCTAGCTAGGCATGTCTTTACTGGAGAAAAAGTTGCAGTGAAAGTGATTGACAAGAGTAAATTGGATGATGTATCAAAGTCCCATTTATTTCAGGAGGTTCGATGTATGAAATTGGTTCAACATCCAAACGTCGTACGCCTATATGAAGTAATAGATACACAAACTAAGCTATATTTAATTTTAGAATTGGGAGATGGTGGTGATTTATATGACTATATCATGAGACACGAATCTGGTCTTTCTGAATCTCTAGCACGTGATTATTTCAGACAAATAGTTCGTGCAATCTCGTATTGCCATAGATTACATGTTGTTCATCGAGACCTCAAACCTGAAAATGTAGTTTTTTTCGAAAAATTAGGACTAGTCAAATTGACTGACTTTGGTTTTAGCAATAAATTTTGTCCAGGTCAGAAATTAGAGACATCCTGTGGTTCACTTGCTTATTCAGCTCCTGAAATATTGTTGGGTGACTCTTATGACGCTCCTGCTGTGGACGTCTGGTCTCTCGGCGTCATTCTCTACATGTTAGTATGTGGACAGGCTCCATTTCAGGAAGCCAATGATAGTGAAACTTTGACAATGATAATGGATTGTAAATATACAATTCCTTCACAAATTTCAAATTCATGTAAAAGACTCATTGGAAGAATGTTAGTGAGAGAACCAGAGAAACGGGCGTGCCTATCCGAAATCGCCACAGATCCTTGGGTCACTGCAGGTGAAGGTGTCACCATAGAAGACTTCGATACGCCGCTCGTTGCCAAAGAACAATTAACTGATGAGGACCGTTCAGCTATAATTCAAAGAATGGTTAGTGGCTCGATAGCCACCAAAGAAGAAATATTGGAAGCTCTGGATAAAAACGAGTACAATCATATAACGGCAACTTATTTTTTACTCGCCGAAAGAAAGCTGAAATCAAATAGGTCGGAAGCGTTGCGTAGACAAGAGGGTCCGCGTCGTCCCGAGGCTCTGAGCGTGCAGCGCGCCGCCCCGCAGTCCTTGCTCGCGCCCCCCGCGCGCGCCCTCGCACCGCGCACGCCACGGACGCCGCAGGACGTCCCACCGAGGTCTAGAAAATGCAGTATAGTAAGAGAAGAAGATGATGACGAGGAGAGCGCGGTCCCGGGCGGTTGCGGTGCGCGGGACGGAGACACGCGACGCGGCTCTCGCTCCGAGGGACGTCTGCATCTCGCAGCCAGGCTCGTGCCGTCCGCGCAGCCCTCCCACCGCCCCGCCCTCTTAGCGGATAACCTCACAAAGGTGAATTTGGATGATGAAGGTGGCGGGATCGCAACGCCGGCACCGAGAGTGCCGCCGCCGGCTCCACCTCCCGCACGCCGGAACCGAATAGAGTGTCGCCGCCGAAGACCTCGCGCCGGCTCTTGCTCCTCGTCTGACTTATCCGATGACGACTCCGAGAAAAAGAAGCGCGCTGCCGAACGAGCCGACGTCGCACTTGATGGTGCTCGCCGAGATTCTCATGACGACTCCAGCGACAGTCAGGAGCGAGGGGCCGGGGCCGGTGCCGGGGCGGTCGGCGGCTCCGAGGCGGGAGCGCAGGCCGCCGGCGACGGCACGTCAGCCGCGGGGGGGCGGGGCGCGAGGGGCGGGGACGCGGGTGGGGACGGCAGGGCGGGCAGCGGGGCGGGCACGGGCCGGCGCCGCCGCACCTCTAAGCATGAGAGTCGGCTCCGCGAGTCGCGGTCGTTGAACCGCATCGCGGAGGTGGAGGAGGCGGGCGGCGCGCGGTGGTCTCGCAGCGGGCTGGTGTCGCTGTGCGGGCGCCCGCTGCTCCGACTGCTGGCGGCGGCGCGTCCCTCTCCGTCAGCGCGCCGGTTGCACGGACTGTGTTAG
Protein
MAVNNNGTSVASGTYDTKIAGLYDLLDTLGSGHFAVVKLARHVFTGEKVAVKVIDKSKLDDVSKSHLFQEVRCMKLVQHPNVVRLYEVIDTQTKLYLILELGDGGDLYDYIMRHESGLSESLARDYFRQIVRAISYCHRLHVVHRDLKPENVVFFEKLGLVKLTDFGFSNKFCPGQKLETSCGSLAYSAPEILLGDSYDAPAVDVWSLGVILYMLVCGQAPFQEANDSETLTMIMDCKYTIPSQISNSCKRLIGRMLVREPEKRACLSEIATDPWVTAGEGVTIEDFDTPLVAKEQLTDEDRSAIIQRMVSGSIATKEEILEALDKNEYNHITATYFLLAERKLKSNRSEALRRQEGPRRPEALSVQRAAPQSLLAPPARALAPRTPRTPQDVPPRSRKCSIVREEDDDEESAVPGGCGARDGDTRRGSRSEGRLHLAARLVPSAQPSHRPALLADNLTKVNLDDEGGGIATPAPRVPPPAPPPARRNRIECRRRRPRAGSCSSSDLSDDDSEKKKRAAERADVALDGARRDSHDDSSDSQERGAGAGAGAVGGSEAGAQAAGDGTSAAGGRGARGGDAGGDGRAGSGAGTGRRRRTSKHESRLRESRSLNRIAEVEEAGGARWSRSGLVSLCGRPLLRLLAAARPSPSARRLHGLC
Summary
Uniprot
H9JQ67
A0A2H1W1L2
A0A3S2M0Z6
A0A194PV79
A0A151X1K1
A0A195CKI0
+ More
A0A158NEY0 A0A195E5G5 F4W452 A0A195B411 A0A026WLQ0 A0A2A3E916 A0A088APN8 A0A310SDM2 A0A0N0U6X1 A0A0L7QT65 A0A154PCB7 A0A2M3ZH88 A0A2M4A954 A0A182NLI6 A0A2M4BAB3 A0A182XV04 A0A2M4BAE5 A0A2M4BA77 W5JJ31 A0A182Q9X1 A0A182V9V6 A0A182LA73 A0A2C9GQ56 Q7PY54 A0A182WSW8 A0A182IT04 A0A182UDP4 A0A1J1I7M9 A0A0K8TZG3 A0A034WG71 W8BYM3 A0A1B0AM47 A0A1A9XPR6
A0A158NEY0 A0A195E5G5 F4W452 A0A195B411 A0A026WLQ0 A0A2A3E916 A0A088APN8 A0A310SDM2 A0A0N0U6X1 A0A0L7QT65 A0A154PCB7 A0A2M3ZH88 A0A2M4A954 A0A182NLI6 A0A2M4BAB3 A0A182XV04 A0A2M4BAE5 A0A2M4BA77 W5JJ31 A0A182Q9X1 A0A182V9V6 A0A182LA73 A0A2C9GQ56 Q7PY54 A0A182WSW8 A0A182IT04 A0A182UDP4 A0A1J1I7M9 A0A0K8TZG3 A0A034WG71 W8BYM3 A0A1B0AM47 A0A1A9XPR6
Pubmed
EMBL
BABH01012067
BABH01012068
BABH01012069
ODYU01005769
SOQ46971.1
RSAL01000080
+ More
RVE48617.1 KQ459590 KPI97306.1 KQ982585 KYQ54279.1 KQ977642 KYN00962.1 ADTU01001494 KQ979609 KYN20142.1 GL887491 EGI71048.1 KQ976625 KYM79007.1 KK107153 QOIP01000012 EZA56977.1 RLU16192.1 KZ288319 PBC28273.1 KQ762927 OAD55257.1 KQ435727 KOX78074.1 KQ414755 KOC61749.1 KQ434870 KZC09535.1 GGFM01007079 MBW27830.1 GGFK01004013 MBW37334.1 GGFJ01000801 MBW49942.1 GGFJ01000831 MBW49972.1 GGFJ01000802 MBW49943.1 ADMH02001085 ETN64146.1 AXCN02000140 AXCN02000141 APCN01000911 AAAB01008987 EAA01241.5 CVRI01000042 CRK95580.1 GDHF01032844 JAI19470.1 GAKP01005630 JAC53322.1 GAMC01002124 GAMC01002123 JAC04433.1 JXJN01000336
RVE48617.1 KQ459590 KPI97306.1 KQ982585 KYQ54279.1 KQ977642 KYN00962.1 ADTU01001494 KQ979609 KYN20142.1 GL887491 EGI71048.1 KQ976625 KYM79007.1 KK107153 QOIP01000012 EZA56977.1 RLU16192.1 KZ288319 PBC28273.1 KQ762927 OAD55257.1 KQ435727 KOX78074.1 KQ414755 KOC61749.1 KQ434870 KZC09535.1 GGFM01007079 MBW27830.1 GGFK01004013 MBW37334.1 GGFJ01000801 MBW49942.1 GGFJ01000831 MBW49972.1 GGFJ01000802 MBW49943.1 ADMH02001085 ETN64146.1 AXCN02000140 AXCN02000141 APCN01000911 AAAB01008987 EAA01241.5 CVRI01000042 CRK95580.1 GDHF01032844 JAI19470.1 GAKP01005630 JAC53322.1 GAMC01002124 GAMC01002123 JAC04433.1 JXJN01000336
Proteomes
UP000005204
UP000283053
UP000053268
UP000075809
UP000078542
UP000005205
+ More
UP000078492 UP000007755 UP000078540 UP000053097 UP000279307 UP000242457 UP000005203 UP000053105 UP000053825 UP000076502 UP000075884 UP000076408 UP000000673 UP000075886 UP000075903 UP000075882 UP000075840 UP000007062 UP000076407 UP000075880 UP000075902 UP000183832 UP000092460 UP000092443
UP000078492 UP000007755 UP000078540 UP000053097 UP000279307 UP000242457 UP000005203 UP000053105 UP000053825 UP000076502 UP000075884 UP000076408 UP000000673 UP000075886 UP000075903 UP000075882 UP000075840 UP000007062 UP000076407 UP000075880 UP000075902 UP000183832 UP000092460 UP000092443
PRIDE
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JQ67
A0A2H1W1L2
A0A3S2M0Z6
A0A194PV79
A0A151X1K1
A0A195CKI0
+ More
A0A158NEY0 A0A195E5G5 F4W452 A0A195B411 A0A026WLQ0 A0A2A3E916 A0A088APN8 A0A310SDM2 A0A0N0U6X1 A0A0L7QT65 A0A154PCB7 A0A2M3ZH88 A0A2M4A954 A0A182NLI6 A0A2M4BAB3 A0A182XV04 A0A2M4BAE5 A0A2M4BA77 W5JJ31 A0A182Q9X1 A0A182V9V6 A0A182LA73 A0A2C9GQ56 Q7PY54 A0A182WSW8 A0A182IT04 A0A182UDP4 A0A1J1I7M9 A0A0K8TZG3 A0A034WG71 W8BYM3 A0A1B0AM47 A0A1A9XPR6
A0A158NEY0 A0A195E5G5 F4W452 A0A195B411 A0A026WLQ0 A0A2A3E916 A0A088APN8 A0A310SDM2 A0A0N0U6X1 A0A0L7QT65 A0A154PCB7 A0A2M3ZH88 A0A2M4A954 A0A182NLI6 A0A2M4BAB3 A0A182XV04 A0A2M4BAE5 A0A2M4BA77 W5JJ31 A0A182Q9X1 A0A182V9V6 A0A182LA73 A0A2C9GQ56 Q7PY54 A0A182WSW8 A0A182IT04 A0A182UDP4 A0A1J1I7M9 A0A0K8TZG3 A0A034WG71 W8BYM3 A0A1B0AM47 A0A1A9XPR6
PDB
5YKS
E-value=4.90872e-151,
Score=1373
Ontologies
GO
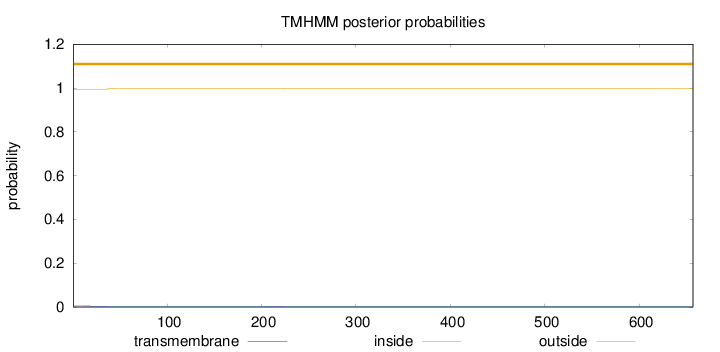
Topology
Length:
657
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.09056
Exp number, first 60 AAs:
0.05954
Total prob of N-in:
0.00532
outside
1 - 657
Population Genetic Test Statistics
Pi
214.060768
Theta
188.094652
Tajima's D
0.572913
CLR
0
CSRT
0.543922803859807
Interpretation
Uncertain
