Pre Gene Modal
BGIBMGA011672
Annotation
PREDICTED:_protein_disulfide-isomerase_TMX3_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.308
Sequence
CDS
ATGAAGTTCATTACGGGATTATACATTATATTTTTTACAAGAATATTTAACGTGTCATCATCTAGAGTTTTAGAACTAAGTGACAGATTTTTAGAGTTTAGCAAAGATGGAATGTGGTTTGTTAAATTCTATGCGCCTTGGTGTTCACACTGTCGGCGCATGGAACCTACTTGGGCGCATGTGGCACAAGCCTTGTATAATACTCCCGTAAAAGTAGCCAAAGTCGATTGTACCCGTTTTACAGCTGTGGCCTCACATTTTCATATCCGAGCTTATCCTACTATCTTGTTTTTAAAAGGATCTTTTCAGCATGAATACACAGGAGATAGGGTTAAAGAGGATTTGATTAATTATGCAATGAGAATGTCACAGCCAGCAGTGCAGAGAATATCTCGTGGTAACAGCATTGAATATTTAAAAGAATCTCATCCTGTATTTTTTGGATTCATTGGCAAACAGGAAGGGCTTTTATGGGAAATGTTTACAATTCATGCTGAGAAATATCAGCCTCATTCCTGGTTTTATGCAATGACCCATGAAGTAATAAAACATGAAATCAAAGTTCCTAATGAAACAGCTATATTTGTTTATAAAGATAATGAAGCATTTTTCTTTGAAGTTAAAGATGAATTACTTGAAGACAAAAATATGTTAAACAACACCTTAGGCAAATGGATTACTTCTGAGAGATTTGGTTTTTTCCCCAAAATAACCCGAGCAAATATTAATAGTCTGATTGATACTGAAAAATATATTGTGATAGTAGTAGTTGCAGAAAATAAACTTGAAGAAATATCACAAACTGAAAGAGATTTTAAAGATATGGTAGAAGGAATTATAATGAAAAGAAAAAATGAGCTACATTCTCGTTTTCAGTTTGGTTGGATGGGTTCACCAGAACTAGCAAATTCTATTGCTATGTCTGAACTTACTGTACCTCATTTATTAGTATTAAACTCAACGACAAGTCATCATCATATTCCAGACGATGATCCAGTATTAATGACACCAGAAGCAATCTCACTGTTTCTTGATCAAATACACAACCAGCAAGCTACCGTATATGGTGGCAATGGCTGGCCAGTAAAACTATATAGAGGTTTCTTTGAAGCAAGAACTACATTGGCAAACATGTGGCATGGAAACCCTGTTCTAACAGCTCTTATCTTTGGTCTGCCACTGGGATTCCTCTCACTGATATGCTATTCAGTGTGTTGTGCTGATATTTTAGATGCTGAAGACGAAGACACACCAGAACTACACGAGAAAAGGGACTAA
Protein
MKFITGLYIIFFTRIFNVSSSRVLELSDRFLEFSKDGMWFVKFYAPWCSHCRRMEPTWAHVAQALYNTPVKVAKVDCTRFTAVASHFHIRAYPTILFLKGSFQHEYTGDRVKEDLINYAMRMSQPAVQRISRGNSIEYLKESHPVFFGFIGKQEGLLWEMFTIHAEKYQPHSWFYAMTHEVIKHEIKVPNETAIFVYKDNEAFFFEVKDELLEDKNMLNNTLGKWITSERFGFFPKITRANINSLIDTEKYIVIVVVAENKLEEISQTERDFKDMVEGIIMKRKNELHSRFQFGWMGSPELANSIAMSELTVPHLLVLNSTTSHHHIPDDDPVLMTPEAISLFLDQIHNQQATVYGGNGWPVKLYRGFFEARTTLANMWHGNPVLTALIFGLPLGFLSLICYSVCCADILDAEDEDTPELHEKRD
Summary
Uniprot
H9JQ66
A0A2H1W1S7
A0A2W1BHX0
A0A2A4JU63
A0A212F401
A0A194Q1Q3
+ More
A0A194QNX5 A0A0L7L181 A0A1L8DUD0 A0A1L8DUG1 A0A182RC15 A0A1Q3FD73 B0WT65 A0A182THR9 A0A182PVV4 A0A182SY05 A0A182VB95 A0A182FHN3 A0A182XCV0 A0A182LJT0 A0A182HM79 A0A182MAR7 A0A182NMC1 W5JI51 Q7Q792 A0A182QHG0 A0A2M4BPU1 A0A2M4BNR3 A0A084VD91 A0A182K7T4 A0A2M4BMZ6 A0A182JL36 A0A2M4AVJ6 A0A182H8I6 A0A1S4FE21 Q175U4 A0A182WLV0 A0A2J7Q711 A0A1B6DVR0 A0A1B6LBI6 A0A1L8DU86 A0A1B6G501 A0A1B6MT86 A0A1B0CKA1 T1HEY8 A0A0T6AVS7 R4WDC3 A0A224XD20 A0A067RD96 D6WGR3 A0A1B6CUT4 E9IZS2 A0A1J1IIL6 A0A1Y1NG91 A0A158NIS9 E2BCN6 A0A0A9XX35 A0A0H3V2Z7 A0A0L7QZB8 A0A2P8YI11 E2ATP4 A0A195CBZ6 A0A0L0CB27 A0A348G611 A0A0A1WTS3 A0A336M0J6 A0A3R7MFR1 B4N406 A0A154PCH6 B3M946 A0A1W4UUP9 W8BMW3 B4PHN0 A0A1I8NHU0 B4LHJ4 A0A0K8WCU0 A0A1I8NHU7 A0A088A7L7 A0A1A9W3B8 A0A034W0S7 A0A0J9RXL5 B4HJ34 Q961B9 B3NI66 B4KW57 A0A0M3QWD0 A0A3L8D2K3 B4J0B1 K7INJ8 A0A1A9YSP7 A0A1B0C360 A0A026W556 E0VGL2 A0A1A9V5V3 A0A1B0G7C7 A0A195BY72 A0A1B0A718 A0A232F3W4 A0A1I8PUX4 A0A1W4X5C7
A0A194QNX5 A0A0L7L181 A0A1L8DUD0 A0A1L8DUG1 A0A182RC15 A0A1Q3FD73 B0WT65 A0A182THR9 A0A182PVV4 A0A182SY05 A0A182VB95 A0A182FHN3 A0A182XCV0 A0A182LJT0 A0A182HM79 A0A182MAR7 A0A182NMC1 W5JI51 Q7Q792 A0A182QHG0 A0A2M4BPU1 A0A2M4BNR3 A0A084VD91 A0A182K7T4 A0A2M4BMZ6 A0A182JL36 A0A2M4AVJ6 A0A182H8I6 A0A1S4FE21 Q175U4 A0A182WLV0 A0A2J7Q711 A0A1B6DVR0 A0A1B6LBI6 A0A1L8DU86 A0A1B6G501 A0A1B6MT86 A0A1B0CKA1 T1HEY8 A0A0T6AVS7 R4WDC3 A0A224XD20 A0A067RD96 D6WGR3 A0A1B6CUT4 E9IZS2 A0A1J1IIL6 A0A1Y1NG91 A0A158NIS9 E2BCN6 A0A0A9XX35 A0A0H3V2Z7 A0A0L7QZB8 A0A2P8YI11 E2ATP4 A0A195CBZ6 A0A0L0CB27 A0A348G611 A0A0A1WTS3 A0A336M0J6 A0A3R7MFR1 B4N406 A0A154PCH6 B3M946 A0A1W4UUP9 W8BMW3 B4PHN0 A0A1I8NHU0 B4LHJ4 A0A0K8WCU0 A0A1I8NHU7 A0A088A7L7 A0A1A9W3B8 A0A034W0S7 A0A0J9RXL5 B4HJ34 Q961B9 B3NI66 B4KW57 A0A0M3QWD0 A0A3L8D2K3 B4J0B1 K7INJ8 A0A1A9YSP7 A0A1B0C360 A0A026W556 E0VGL2 A0A1A9V5V3 A0A1B0G7C7 A0A195BY72 A0A1B0A718 A0A232F3W4 A0A1I8PUX4 A0A1W4X5C7
Pubmed
19121390
28756777
22118469
26354079
26227816
20966253
+ More
20920257 23761445 12364791 14747013 17210077 24438588 26483478 17510324 23691247 24845553 18362917 19820115 21282665 28004739 21347285 20798317 25401762 26823975 29403074 26108605 25830018 17994087 24495485 17550304 25315136 25348373 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 20075255 24508170 20566863 28648823
20920257 23761445 12364791 14747013 17210077 24438588 26483478 17510324 23691247 24845553 18362917 19820115 21282665 28004739 21347285 20798317 25401762 26823975 29403074 26108605 25830018 17994087 24495485 17550304 25315136 25348373 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 30249741 20075255 24508170 20566863 28648823
EMBL
BABH01012070
ODYU01005769
SOQ46973.1
KZ150049
PZC74448.1
NWSH01000661
+ More
PCG74950.1 AGBW02010462 OWR48460.1 KQ459590 KPI97330.1 KQ461190 KPJ07217.1 JTDY01003618 KOB69253.1 GFDF01004047 JAV10037.1 GFDF01004048 JAV10036.1 GFDL01009555 JAV25490.1 DS232080 EDS34208.1 APCN01004377 AXCM01001426 ADMH02001337 ETN62963.1 AAAB01008960 EAA11944.3 AXCN02000935 GGFJ01005657 MBW54798.1 GGFJ01005576 MBW54717.1 ATLV01011192 KE524650 KFB35935.1 GGFJ01005007 MBW54148.1 GGFK01011498 MBW44819.1 JXUM01029021 JXUM01029022 KQ560839 KXJ80665.1 CH477395 EAT41861.1 NEVH01017447 PNF24377.1 GEDC01007532 JAS29766.1 GEBQ01018914 JAT21063.1 GFDF01004097 JAV09987.1 GECZ01012244 JAS57525.1 GEBQ01000827 JAT39150.1 AJWK01015937 AJWK01015938 AJWK01015939 ACPB03013538 ACPB03013539 LJIG01022694 KRT79210.1 AK417548 BAN20763.1 GFTR01006080 JAW10346.1 KK852777 KDR16743.1 KQ971327 EEZ99618.1 GEDC01020167 JAS17131.1 GL767232 EFZ13968.1 CVRI01000054 CRL00032.1 GEZM01003080 GEZM01003079 JAV96932.1 ADTU01017117 GL447336 EFN86546.1 GBHO01020176 GDHC01000604 JAG23428.1 JAQ18025.1 KJ939606 AJE70277.1 KQ414681 KOC63943.1 PYGN01000579 PSN43899.1 GL442637 EFN63198.1 KQ978068 KYM97613.1 JRES01000645 KNC29623.1 FX985550 BBF97884.1 GBXI01012382 JAD01910.1 UFQT01000289 SSX22831.1 QCYY01001116 ROT80473.1 CH964095 EDW79361.1 KQ434870 KZC09487.1 CH902618 EDV40030.1 GAMC01006428 JAC00128.1 CM000159 EDW95468.1 CH940647 EDW68524.1 GDHF01003412 JAI48902.1 GAKP01010678 JAC48274.1 CM002912 KMY99969.1 CH480815 EDW41690.1 AE014296 AY051696 AAF49525.3 AAK93120.1 CH954178 EDV52222.1 CH933809 EDW17445.1 CP012525 ALC43933.1 QOIP01000014 RLU14737.1 CH916366 EDV95712.1 JXJN01024845 KK107405 EZA51207.1 DS235149 EEB12518.1 CCAG010006833 KQ976394 KYM93250.1 NNAY01001103 OXU25133.1
PCG74950.1 AGBW02010462 OWR48460.1 KQ459590 KPI97330.1 KQ461190 KPJ07217.1 JTDY01003618 KOB69253.1 GFDF01004047 JAV10037.1 GFDF01004048 JAV10036.1 GFDL01009555 JAV25490.1 DS232080 EDS34208.1 APCN01004377 AXCM01001426 ADMH02001337 ETN62963.1 AAAB01008960 EAA11944.3 AXCN02000935 GGFJ01005657 MBW54798.1 GGFJ01005576 MBW54717.1 ATLV01011192 KE524650 KFB35935.1 GGFJ01005007 MBW54148.1 GGFK01011498 MBW44819.1 JXUM01029021 JXUM01029022 KQ560839 KXJ80665.1 CH477395 EAT41861.1 NEVH01017447 PNF24377.1 GEDC01007532 JAS29766.1 GEBQ01018914 JAT21063.1 GFDF01004097 JAV09987.1 GECZ01012244 JAS57525.1 GEBQ01000827 JAT39150.1 AJWK01015937 AJWK01015938 AJWK01015939 ACPB03013538 ACPB03013539 LJIG01022694 KRT79210.1 AK417548 BAN20763.1 GFTR01006080 JAW10346.1 KK852777 KDR16743.1 KQ971327 EEZ99618.1 GEDC01020167 JAS17131.1 GL767232 EFZ13968.1 CVRI01000054 CRL00032.1 GEZM01003080 GEZM01003079 JAV96932.1 ADTU01017117 GL447336 EFN86546.1 GBHO01020176 GDHC01000604 JAG23428.1 JAQ18025.1 KJ939606 AJE70277.1 KQ414681 KOC63943.1 PYGN01000579 PSN43899.1 GL442637 EFN63198.1 KQ978068 KYM97613.1 JRES01000645 KNC29623.1 FX985550 BBF97884.1 GBXI01012382 JAD01910.1 UFQT01000289 SSX22831.1 QCYY01001116 ROT80473.1 CH964095 EDW79361.1 KQ434870 KZC09487.1 CH902618 EDV40030.1 GAMC01006428 JAC00128.1 CM000159 EDW95468.1 CH940647 EDW68524.1 GDHF01003412 JAI48902.1 GAKP01010678 JAC48274.1 CM002912 KMY99969.1 CH480815 EDW41690.1 AE014296 AY051696 AAF49525.3 AAK93120.1 CH954178 EDV52222.1 CH933809 EDW17445.1 CP012525 ALC43933.1 QOIP01000014 RLU14737.1 CH916366 EDV95712.1 JXJN01024845 KK107405 EZA51207.1 DS235149 EEB12518.1 CCAG010006833 KQ976394 KYM93250.1 NNAY01001103 OXU25133.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000075900 UP000002320 UP000075902 UP000075885 UP000075901 UP000075903 UP000069272 UP000076407 UP000075882 UP000075840 UP000075883 UP000075884 UP000000673 UP000007062 UP000075886 UP000030765 UP000075881 UP000075880 UP000069940 UP000249989 UP000008820 UP000075920 UP000235965 UP000092461 UP000015103 UP000027135 UP000007266 UP000183832 UP000005205 UP000008237 UP000053825 UP000245037 UP000000311 UP000078542 UP000037069 UP000283509 UP000007798 UP000076502 UP000007801 UP000192221 UP000002282 UP000095301 UP000008792 UP000005203 UP000091820 UP000001292 UP000000803 UP000008711 UP000009192 UP000092553 UP000279307 UP000001070 UP000002358 UP000092443 UP000092460 UP000053097 UP000009046 UP000078200 UP000092444 UP000078540 UP000092445 UP000215335 UP000095300 UP000192223
UP000075900 UP000002320 UP000075902 UP000075885 UP000075901 UP000075903 UP000069272 UP000076407 UP000075882 UP000075840 UP000075883 UP000075884 UP000000673 UP000007062 UP000075886 UP000030765 UP000075881 UP000075880 UP000069940 UP000249989 UP000008820 UP000075920 UP000235965 UP000092461 UP000015103 UP000027135 UP000007266 UP000183832 UP000005205 UP000008237 UP000053825 UP000245037 UP000000311 UP000078542 UP000037069 UP000283509 UP000007798 UP000076502 UP000007801 UP000192221 UP000002282 UP000095301 UP000008792 UP000005203 UP000091820 UP000001292 UP000000803 UP000008711 UP000009192 UP000092553 UP000279307 UP000001070 UP000002358 UP000092443 UP000092460 UP000053097 UP000009046 UP000078200 UP000092444 UP000078540 UP000092445 UP000215335 UP000095300 UP000192223
Pfam
PF00085 Thioredoxin
SUPFAM
SSF52833
SSF52833
ProteinModelPortal
H9JQ66
A0A2H1W1S7
A0A2W1BHX0
A0A2A4JU63
A0A212F401
A0A194Q1Q3
+ More
A0A194QNX5 A0A0L7L181 A0A1L8DUD0 A0A1L8DUG1 A0A182RC15 A0A1Q3FD73 B0WT65 A0A182THR9 A0A182PVV4 A0A182SY05 A0A182VB95 A0A182FHN3 A0A182XCV0 A0A182LJT0 A0A182HM79 A0A182MAR7 A0A182NMC1 W5JI51 Q7Q792 A0A182QHG0 A0A2M4BPU1 A0A2M4BNR3 A0A084VD91 A0A182K7T4 A0A2M4BMZ6 A0A182JL36 A0A2M4AVJ6 A0A182H8I6 A0A1S4FE21 Q175U4 A0A182WLV0 A0A2J7Q711 A0A1B6DVR0 A0A1B6LBI6 A0A1L8DU86 A0A1B6G501 A0A1B6MT86 A0A1B0CKA1 T1HEY8 A0A0T6AVS7 R4WDC3 A0A224XD20 A0A067RD96 D6WGR3 A0A1B6CUT4 E9IZS2 A0A1J1IIL6 A0A1Y1NG91 A0A158NIS9 E2BCN6 A0A0A9XX35 A0A0H3V2Z7 A0A0L7QZB8 A0A2P8YI11 E2ATP4 A0A195CBZ6 A0A0L0CB27 A0A348G611 A0A0A1WTS3 A0A336M0J6 A0A3R7MFR1 B4N406 A0A154PCH6 B3M946 A0A1W4UUP9 W8BMW3 B4PHN0 A0A1I8NHU0 B4LHJ4 A0A0K8WCU0 A0A1I8NHU7 A0A088A7L7 A0A1A9W3B8 A0A034W0S7 A0A0J9RXL5 B4HJ34 Q961B9 B3NI66 B4KW57 A0A0M3QWD0 A0A3L8D2K3 B4J0B1 K7INJ8 A0A1A9YSP7 A0A1B0C360 A0A026W556 E0VGL2 A0A1A9V5V3 A0A1B0G7C7 A0A195BY72 A0A1B0A718 A0A232F3W4 A0A1I8PUX4 A0A1W4X5C7
A0A194QNX5 A0A0L7L181 A0A1L8DUD0 A0A1L8DUG1 A0A182RC15 A0A1Q3FD73 B0WT65 A0A182THR9 A0A182PVV4 A0A182SY05 A0A182VB95 A0A182FHN3 A0A182XCV0 A0A182LJT0 A0A182HM79 A0A182MAR7 A0A182NMC1 W5JI51 Q7Q792 A0A182QHG0 A0A2M4BPU1 A0A2M4BNR3 A0A084VD91 A0A182K7T4 A0A2M4BMZ6 A0A182JL36 A0A2M4AVJ6 A0A182H8I6 A0A1S4FE21 Q175U4 A0A182WLV0 A0A2J7Q711 A0A1B6DVR0 A0A1B6LBI6 A0A1L8DU86 A0A1B6G501 A0A1B6MT86 A0A1B0CKA1 T1HEY8 A0A0T6AVS7 R4WDC3 A0A224XD20 A0A067RD96 D6WGR3 A0A1B6CUT4 E9IZS2 A0A1J1IIL6 A0A1Y1NG91 A0A158NIS9 E2BCN6 A0A0A9XX35 A0A0H3V2Z7 A0A0L7QZB8 A0A2P8YI11 E2ATP4 A0A195CBZ6 A0A0L0CB27 A0A348G611 A0A0A1WTS3 A0A336M0J6 A0A3R7MFR1 B4N406 A0A154PCH6 B3M946 A0A1W4UUP9 W8BMW3 B4PHN0 A0A1I8NHU0 B4LHJ4 A0A0K8WCU0 A0A1I8NHU7 A0A088A7L7 A0A1A9W3B8 A0A034W0S7 A0A0J9RXL5 B4HJ34 Q961B9 B3NI66 B4KW57 A0A0M3QWD0 A0A3L8D2K3 B4J0B1 K7INJ8 A0A1A9YSP7 A0A1B0C360 A0A026W556 E0VGL2 A0A1A9V5V3 A0A1B0G7C7 A0A195BY72 A0A1B0A718 A0A232F3W4 A0A1I8PUX4 A0A1W4X5C7
PDB
1X5D
E-value=3.47892e-16,
Score=208
Ontologies
GO
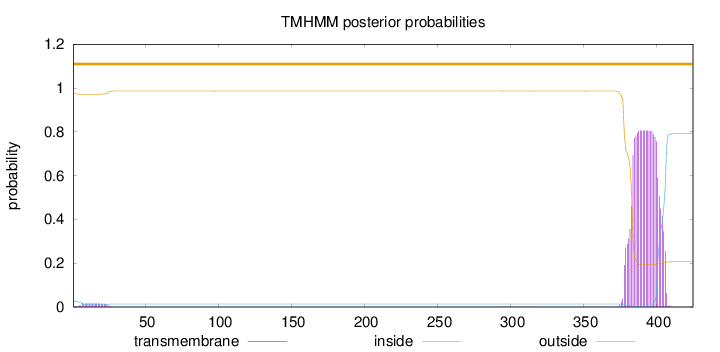
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.638737
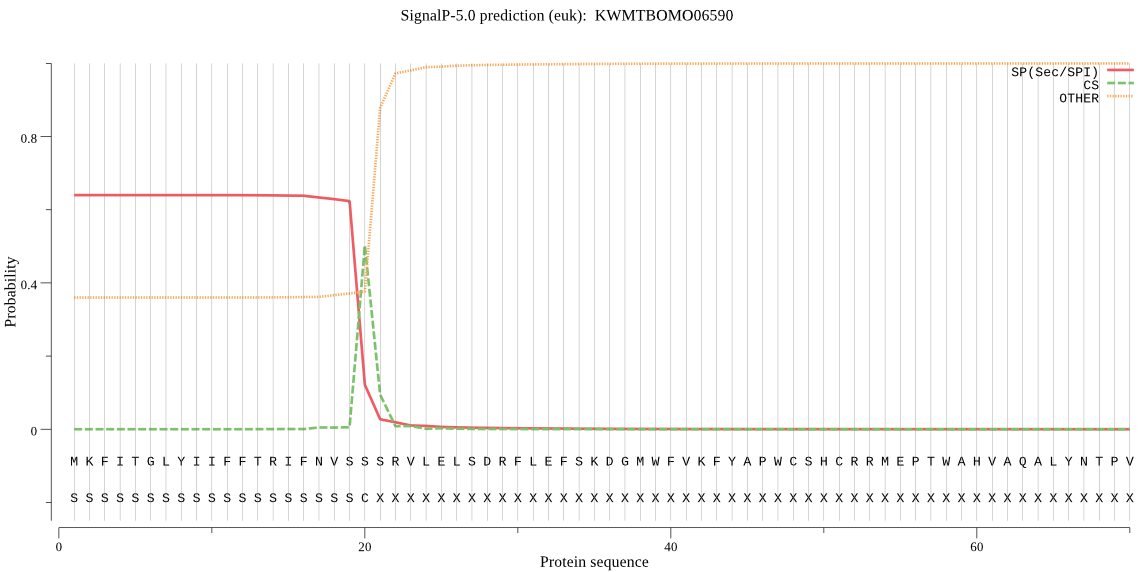
Length:
425
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
18.35379
Exp number, first 60 AAs:
0.3136
Total prob of N-in:
0.02620
outside
1 - 425
Population Genetic Test Statistics
Pi
226.380457
Theta
210.219127
Tajima's D
0.206283
CLR
0.033531
CSRT
0.421828908554572
Interpretation
Uncertain
