Gene
KWMTBOMO06586
Pre Gene Modal
BGIBMGA011807
Annotation
PREDICTED:_potassium_voltage-gated_channel_protein_Shab_isoform_X3_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.144
Sequence
CDS
ATGGGAGGCGAGGGGGCGGGCCGCGGCGCCGCGCGCTGTCACGTGGGCGACTCGCCCCCGCGCGCGCCCGCCATGGGGTTCGCGGCGCCTGCGCAGGACGCGCTGCTCGACCGCGGGCTCGCACCTGCACCCGCGCCTGCACCCACACTCGCCGCGCTGCACCGGGCGCATTCCAGGTCGATGTCGTCTTTACCTCCGGAGCCGTTTATGATGCTCCGTGCTAAAGCTTTAAGCAAGCGCGTCATCATCAACGTGGGTGGTGTAAGACACGAGGTGCTATGGAGAACCCTAGAACGACTGCCGCACACGCGGCTTGGCCGACTACGGGACTGCAACACGCACGAAGCCATCACAGAACTCTGCGATGACTATTCGCTTCAGGATAACGAGTACTTTTTTGACAGACACCCGAAGAGCTTTAGTTCGATTTTAAATTTCTATCGAACTGGTAAACTTCACTTAGTGGATGAAATGTGTGTGTTGGCATTCAGCGATGATCTCGAGTATTGGGGCGTGGATGAGTTGTATCTGGAGTCTTGTTGTCAACACAAGTATCACCAACGGAAGGAGCACGTTCATGAGGAGATGCGCAAGGAAGCCGAAAGTTTAAGGCAGCGGGAAGAAGAGGAGTTTGGTCAGGGCACATGTGCTCAATATCAGAAATGGCTTTGGGACTTATTGGAGAAGCCGACCACGAGCATTGCGGCGCGGGTGATTGCTGTGATATCGATTCTTTTCATCGTGCTCTCCACGATAGGGCTCACTCTCAACACAATCCCTCAAATGCAAGTTCACGATGACAAAGGCCCGATGGACAATCCGAAGCTGGCCATGGTCGAGGCCGTGTGCATCACTTGGTTCACGCTCGAGTATGTGCTGCGGTTTAGTGCCTCACCGGATAAGTGGAAGTTTTTTAAAGGTGGACTGAACGTTATAGACTTACTGGCGATACTGCCGTATTTTGTGTCTCTATTCCTTCTCGAAACAAATAAGAACGCTACGGATCAGTTCCAAGACGTGCGTCGTGTTGTTCAAGTATTTCGTATAATGCGAATACTGCGCATCCTCAAACTTGCACGACATTCGACCGGGTTGCAATCCCTCGGTTTCACACTCAGGAACTCTTATAAAGAGCTCGGCCTTCTAATGTTGTTCCTCGCTATGGGAGTGCTTATATTTTCTTCGCTGGCGTACTTTGCCGAAAAGGAAGAACCTGGTACCAAATTCATTTCGATTCCGGAAACATTCTGGTGGGCCGGCATCACCATGACGACAGTCGGATACGGTGACATTTACCCTACGACTCCGTTGGGCAAGGTCATCGGCTCAGTATGCTGTATCTGCGGCGTGTTGGTAATTGCGTTACCCATTCCCATTATCGTCAACAATTTCGCCGAGTTTTACAAGAATCAGATGCGGCGCGAGAAAGCCCTTAAGCGTCGAGAGGCTCTCGAGCGAGCCAAGCGCGAGGGCTCCATAGTATCTTTCCACCACATAAACCTACGAGACGCTTTCGCCAAGAGCATGGACCTCATCGACGTCATTGTTGACACAGGACACAATTTGTCGCAGGCGGATGCTACTAGTTTGGAAGGCGAAGCTCCTGTGACCACTGGCTGCTATAAACAATACGAACACAGTATACCCAACAAGCTCTCTCTCGATACAGAATCAAATCAGGGTAGCCCTAAAAAAGAGAGACGGTACTCCAATGAAGTCCCCGGCACGCCAGAGAAGCGTCTCCTCATAAACCCCGAGCCTACTCCTGGTTACAAGGAGTTCATAGATGCAGAGCAATTGATGCCTATGTCCACGTCTGATCTGCGCCAGCCAGTCTGTGTAGAACTGGCTTGTCTTCAGCGACATCCTGCACCCAAGGCTCTACCAGAGGCCACACCTAATAGTTTAGATAGCCCAGATACATTTGCATCATGCCTTACGCACCCTTTTCCTTCAGAGGGAGACATCGCATGCGAGAACGACTCGTCAAACTTGTATGTGAATCCTCTGGATGGTAGCGGCGGGGGCGGCGCGGGTGCGGGAGAGGGTGGGGTACCGGGGGTACGGCGGAGCGCTTCCGGGGACGGGAACGTGTTGTGTGCGAAGGGTTCGCAGGGCGAGGTGTCACCGCGCGGGTCGCGGGCCAGTCTGAGCGGCGCTGTAGTGCGGCCCCTCGACGCCTCGAGGACCAGTCTGCCGCAGCCCTTAGACGATTTACCCGACGACACAAAGCCCAAACACAGAAAAGCTCGCTTCCAACAGGAATGGACCGACGAAGAGAGGAAGCCGCCGAGGGGTATAGCAACGAGGATAATCAATCACCACATCCTTTTTGGTAAAAACGGCAACAAACGGGGGGGCGGCTGGCCGTCAGAAGGCGACTCGCGTTCCCGGTCCATCCTCAAGAACAAGGAGTCGCGCGGTACAGACAGCGAGCTGCTTCTGCAAGAAACTACATCACTCGGCGCAAATGGTGACTCCGGTAGTGAATCATCGCCGATCCGACTAAAAGACTTCAAAGGCCAAAACCACAAAGGTGGAGCACCGGAGAGGGATAACGTGATGACGTAG
Protein
MGGEGAGRGAARCHVGDSPPRAPAMGFAAPAQDALLDRGLAPAPAPAPTLAALHRAHSRSMSSLPPEPFMMLRAKALSKRVIINVGGVRHEVLWRTLERLPHTRLGRLRDCNTHEAITELCDDYSLQDNEYFFDRHPKSFSSILNFYRTGKLHLVDEMCVLAFSDDLEYWGVDELYLESCCQHKYHQRKEHVHEEMRKEAESLRQREEEEFGQGTCAQYQKWLWDLLEKPTTSIAARVIAVISILFIVLSTIGLTLNTIPQMQVHDDKGPMDNPKLAMVEAVCITWFTLEYVLRFSASPDKWKFFKGGLNVIDLLAILPYFVSLFLLETNKNATDQFQDVRRVVQVFRIMRILRILKLARHSTGLQSLGFTLRNSYKELGLLMLFLAMGVLIFSSLAYFAEKEEPGTKFISIPETFWWAGITMTTVGYGDIYPTTPLGKVIGSVCCICGVLVIALPIPIIVNNFAEFYKNQMRREKALKRREALERAKREGSIVSFHHINLRDAFAKSMDLIDVIVDTGHNLSQADATSLEGEAPVTTGCYKQYEHSIPNKLSLDTESNQGSPKKERRYSNEVPGTPEKRLLINPEPTPGYKEFIDAEQLMPMSTSDLRQPVCVELACLQRHPAPKALPEATPNSLDSPDTFASCLTHPFPSEGDIACENDSSNLYVNPLDGSGGGGAGAGEGGVPGVRRSASGDGNVLCAKGSQGEVSPRGSRASLSGAVVRPLDASRTSLPQPLDDLPDDTKPKHRKARFQQEWTDEERKPPRGIATRIINHHILFGKNGNKRGGGWPSEGDSRSRSILKNKESRGTDSELLLQETTSLGANGDSGSESSPIRLKDFKGQNHKGGAPERDNVMT
Summary
Similarity
Belongs to the potassium channel family.
Uniprot
EMBL
BABH01012076
KQ459590
KPI97310.1
RSAL01000209
RVE44242.1
KZ150049
+ More
PZC74452.1 AGBW02014142 OWR42198.1 ODYU01007483 SOQ50291.1 JTDY01004183 KOB68489.1 KQ461190 KPJ07233.1 GEDC01012792 JAS24506.1 QOIP01000010 RLU17854.1 ADTU01002821 ADTU01002822 ADTU01002823 ADTU01002824 ADTU01002825 ADTU01002826 KQ434908 KZC11316.1 GECU01006892 JAT00815.1 GBYB01003868 JAG73635.1 ACPB03004531 ACPB03004532 ABLF02035646 ABLF02035648 DS232049 EDS33126.1 CH477597 EAT38515.1
PZC74452.1 AGBW02014142 OWR42198.1 ODYU01007483 SOQ50291.1 JTDY01004183 KOB68489.1 KQ461190 KPJ07233.1 GEDC01012792 JAS24506.1 QOIP01000010 RLU17854.1 ADTU01002821 ADTU01002822 ADTU01002823 ADTU01002824 ADTU01002825 ADTU01002826 KQ434908 KZC11316.1 GECU01006892 JAT00815.1 GBYB01003868 JAG73635.1 ACPB03004531 ACPB03004532 ABLF02035646 ABLF02035648 DS232049 EDS33126.1 CH477597 EAT38515.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF54695
SSF54695
Gene 3D
ProteinModelPortal
PDB
4JTD
E-value=2.72532e-62,
Score=608
Ontologies
GO
PANTHER
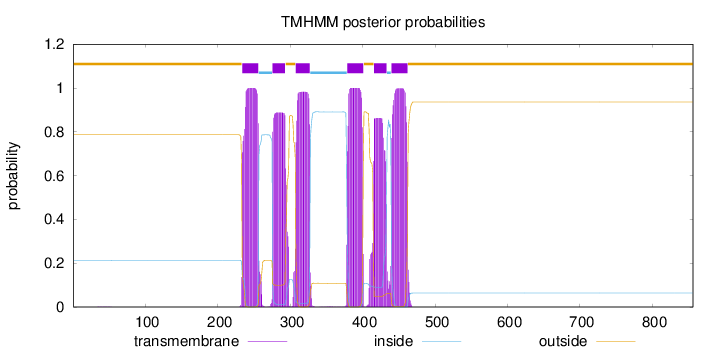
Topology
Length:
856
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
122.01564
Exp number, first 60 AAs:
0.00532
Total prob of N-in:
0.21313
outside
1 - 233
TMhelix
234 - 256
inside
257 - 275
TMhelix
276 - 293
outside
294 - 307
TMhelix
308 - 327
inside
328 - 378
TMhelix
379 - 401
outside
402 - 415
TMhelix
416 - 433
inside
434 - 439
TMhelix
440 - 462
outside
463 - 856
Population Genetic Test Statistics
Pi
224.396591
Theta
182.410245
Tajima's D
0.401618
CLR
0.485621
CSRT
0.486125693715314
Interpretation
Uncertain
