Gene
KWMTBOMO06585
Pre Gene Modal
BGIBMGA011669
Annotation
PREDICTED:_N-alpha-acetyltransferase_60_[Bombyx_mori]
Full name
N-alpha-acetyltransferase 60
Alternative Name
NatF catalytic subunit
Location in the cell
PlasmaMembrane Reliability : 3.824
Sequence
CDS
ATGGCTGGGTTTAGCTGGTATCTGTCTGAAGGCTTCCAAGTTATAATTGAGAAGTCTAAAGACAAGAAATGCTCTCTAAAAGATGTACAATTACGTTTCTTGTGCCCTGATGATTTAGAAGAGGTTAGGTCTCTGTGCAGAGACTGGTTTCCTATCGAATACCCGCAGTCTTGGTATGAAGACATCACATCCTCGGACAGGTTCTTCGCGCTCGCGGCAGTGTACAAGAACCAGATCATAGGTCTCATTGTTGCCGAAATAAAACCTTATTTAAAACTGAATGCCGAAGACAGAGGTATCTTGTCGAGATGGTTCGCTTCCAAGGATACATTAGTTGCTTATATATTGTCATTGGGAGTGGTGCGTTCGCACCGTCGTTCAGGGGTGGCAACGATGCTGCTCGATGTTCTGATCAACCACCTGTCGGGGCCGGTGCCCCAGCCGCCGCACGAACATCGAGTCAAGGCTATATTTCTGCACGTCCTCACGACCAACAACGAGGCTATACTGTTTTATGAACACAAAAGATTCCTTCTACACTCGTTTCTTCCGTACTACTACTCGATAAAGGGCCGTTGTAAAGACGGTTTCACATACGTGTACTATGTGAACGGGGGCCACGCGCCGTGGAGCCTCATCGACTACATGAAGTTCATGGCTCGGACAGCGTGGAGGGGCGGAGGCCTATATCCGTGGCTGTGGGGGAAACTGAGAGCGGCGCTCACTATAGCGTGGCACAGTTTTACGTCGTCTTCGAATACTGTAAAATTTAGGGGTGCGGTATACTGA
Protein
MAGFSWYLSEGFQVIIEKSKDKKCSLKDVQLRFLCPDDLEEVRSLCRDWFPIEYPQSWYEDITSSDRFFALAAVYKNQIIGLIVAEIKPYLKLNAEDRGILSRWFASKDTLVAYILSLGVVRSHRRSGVATMLLDVLINHLSGPVPQPPHEHRVKAIFLHVLTTNNEAILFYEHKRFLLHSFLPYYYSIKGRCKDGFTYVYYVNGGHAPWSLIDYMKFMARTAWRGGGLYPWLWGKLRAALTIAWHSFTSSSNTVKFRGAVY
Summary
Description
Displays alpha (N-terminal) acetyltransferase activity towards a range of N-terminal sequences including those starting with Met-Lys, Met-Val, Met-Ala and Met-Met. Required for normal chromosomal segregation during anaphase.
Isoform A: Shows histone acetyltransferase activity toward free histones.
Isoform B: Does not show histone acetyltransferase activity toward free histones.
Isoform A: Shows histone acetyltransferase activity toward free histones.
Isoform B: Does not show histone acetyltransferase activity toward free histones.
Catalytic Activity
acetyl-CoA + N-terminal L-methionyl-[transmembrane protein] = CoA + H(+) + N-terminal N(alpha)-acetyl-L-methionyl-[transmembrane protein]
acetyl-CoA + L-lysyl-[protein] = CoA + H(+) + N(6)-acetyl-L-lysyl-[protein]
acetyl-CoA + L-lysyl-[protein] = CoA + H(+) + N(6)-acetyl-L-lysyl-[protein]
Similarity
Belongs to the acetyltransferase family. NAA60 subfamily.
Keywords
Acyltransferase
Alternative splicing
Chromatin regulator
Chromosome partition
Complete proteome
Reference proteome
Transferase
Feature
chain N-alpha-acetyltransferase 60
splice variant In isoform B and isoform C.
splice variant In isoform B and isoform C.
Uniprot
H9JQ63
A0A2W1BRR9
A0A2H1VSZ3
A0A212EL51
A0A194QNZ0
A0A194PV85
+ More
A0A0L7KYZ1 A0A1Y1L6H4 A0A139WK49 A0A0P6J6A3 A0A1Q3G351 A0A182JG47 A0A182XEX5 A0A1Y1L305 A0A2M3ZB36 A0A2M4BXH6 A0A2M4AU45 A0A2M4CKK6 A0A2M4CKE3 A0A2M4CKC5 A0A1Y9INA8 A0A1L8DCV4 A0A182H3N9 A0A023EJQ4 A0A067QKR8 U5EL81 A0A0J9RTX3 Q95SX8-2 B3NGL1 B4HLA1 A0A0K8TKZ5 B4PEC9 A0A1Y1L7W5 D6WGD7 A0A0A9XY07 B3MA74 A0A1W4U9U8 B4L0K6 A0A182T5X9 E2C870 A0A1S4GND8 B4LGA7 A0A1I8NF75 A0A0L7QRE9 A0A151X239 A0A2M4DSE6 B4J2P6 A0A195EU61 A0A182RX08 A0A2M3ZJ68 F4W499 A0A195B3J0 B4N4A1 A0A158NFW6 A0A151INW9 E9IE22 A0A182M1M3 A0A182N1Q6 A0A1B6BXA0 A0A026VUY1 B5DQ59 B4HAF6 A0A1I8QAL0 A0A0M9AB05 A0A2A3EM93 V9II72 A0A088AAN9 Q7PQ00 K7IR38 T1GN36 A0A1B6LDQ0 A0A182HP01 A0A1B0D7I0 A0A1B6IIX5 A0A1B6GW07 B4QNN5 A0A1Y1L5A3 Q95SX8 A0A224XW67 A0A069DRG7 A0A0J9UIB6 A0A0Q5U3R3 A0A0K8V4V0 A0A034WBA5 A0A195E5M6 E2A733 A0A0R1DUX1 Q95SX8-3 A0A0P8YG23 A0A0A1WUK0 A0A1W4UL81 A0A1A9YEB9 A0A1B0AVM4 A0A0Q9XLY8 A0A0T6ASX3 A0A0K8VZU5 A0A0Q9WJ68 B0WH57 J3JVU9 A0A0A1X4X6
A0A0L7KYZ1 A0A1Y1L6H4 A0A139WK49 A0A0P6J6A3 A0A1Q3G351 A0A182JG47 A0A182XEX5 A0A1Y1L305 A0A2M3ZB36 A0A2M4BXH6 A0A2M4AU45 A0A2M4CKK6 A0A2M4CKE3 A0A2M4CKC5 A0A1Y9INA8 A0A1L8DCV4 A0A182H3N9 A0A023EJQ4 A0A067QKR8 U5EL81 A0A0J9RTX3 Q95SX8-2 B3NGL1 B4HLA1 A0A0K8TKZ5 B4PEC9 A0A1Y1L7W5 D6WGD7 A0A0A9XY07 B3MA74 A0A1W4U9U8 B4L0K6 A0A182T5X9 E2C870 A0A1S4GND8 B4LGA7 A0A1I8NF75 A0A0L7QRE9 A0A151X239 A0A2M4DSE6 B4J2P6 A0A195EU61 A0A182RX08 A0A2M3ZJ68 F4W499 A0A195B3J0 B4N4A1 A0A158NFW6 A0A151INW9 E9IE22 A0A182M1M3 A0A182N1Q6 A0A1B6BXA0 A0A026VUY1 B5DQ59 B4HAF6 A0A1I8QAL0 A0A0M9AB05 A0A2A3EM93 V9II72 A0A088AAN9 Q7PQ00 K7IR38 T1GN36 A0A1B6LDQ0 A0A182HP01 A0A1B0D7I0 A0A1B6IIX5 A0A1B6GW07 B4QNN5 A0A1Y1L5A3 Q95SX8 A0A224XW67 A0A069DRG7 A0A0J9UIB6 A0A0Q5U3R3 A0A0K8V4V0 A0A034WBA5 A0A195E5M6 E2A733 A0A0R1DUX1 Q95SX8-3 A0A0P8YG23 A0A0A1WUK0 A0A1W4UL81 A0A1A9YEB9 A0A1B0AVM4 A0A0Q9XLY8 A0A0T6ASX3 A0A0K8VZU5 A0A0Q9WJ68 B0WH57 J3JVU9 A0A0A1X4X6
EC Number
2.3.1.259
2.3.1.48
2.3.1.48
Pubmed
19121390
28756777
22118469
26354079
26227816
28004739
+ More
18362917 19820115 26999592 26483478 24945155 24845553 22936249 10731132 12537572 12537569 21981917 21750686 17994087 26369729 17550304 25401762 26823975 20798317 12364791 25315136 21719571 21347285 21282665 24508170 30249741 15632085 20075255 26334808 25348373 25830018 22516182
18362917 19820115 26999592 26483478 24945155 24845553 22936249 10731132 12537572 12537569 21981917 21750686 17994087 26369729 17550304 25401762 26823975 20798317 12364791 25315136 21719571 21347285 21282665 24508170 30249741 15632085 20075255 26334808 25348373 25830018 22516182
EMBL
BABH01012076
KZ150049
PZC74453.1
ODYU01004271
SOQ43965.1
AGBW02014142
+ More
OWR42197.1 KQ461190 KPJ07232.1 KQ459590 KPI97311.1 JTDY01004183 KOB68488.1 GEZM01068121 JAV67166.1 KQ971329 KYB28350.1 GDUN01000180 JAN95739.1 GFDL01000876 JAV34169.1 GEZM01068118 JAV67168.1 GGFM01004972 MBW25723.1 GGFJ01008636 MBW57777.1 GGFK01010998 MBW44319.1 GGFL01001597 MBW65775.1 GGFL01001595 MBW65773.1 GGFL01001596 MBW65774.1 GFDF01009940 JAV04144.1 JXUM01107731 KQ565169 KXJ71257.1 GAPW01004312 JAC09286.1 KK853239 KDR09509.1 GANO01001522 JAB58349.1 CM002912 KMY98744.1 AE014296 AY060437 BT001491 AAL25476.1 AAS65049.1 CH954178 EDV51247.1 CH480815 EDW40920.1 GDAI01002770 JAI14833.1 CM000159 EDW92967.1 GEZM01068119 JAV67167.1 EFA01153.1 GBHO01021469 GBRD01010193 GDHC01010432 GDHC01008823 GDHC01000313 JAG22135.1 JAG55631.1 JAQ08197.1 JAQ09806.1 JAQ18316.1 CH902618 EDV39088.1 CH933809 EDW18083.1 GL453568 EFN75873.1 AAAB01008900 CH940647 EDW69415.1 KQ414784 KOC61129.1 KQ982585 KYQ54326.1 GGFL01016299 MBW80477.1 CH916366 EDV96037.1 KQ981979 KYN31442.1 GGFM01007737 MBW28488.1 GL887491 EGI71095.1 KQ976625 KYM79058.1 CH964095 EDW78975.1 ADTU01001538 KQ976932 KYN07038.1 GL762556 EFZ21140.1 AXCM01001477 GEDC01031410 JAS05888.1 KK107847 QOIP01000005 EZA47485.1 RLU23041.1 CH379069 EDY73726.1 CH479240 EDW37554.1 KQ435711 KOX79479.1 KZ288210 PBC32923.1 JR046405 AEY60186.1 EAA09480.5 CAQQ02182124 CAQQ02182125 GEBQ01018162 JAT21815.1 APCN01002823 AJVK01026926 GECU01020845 JAS86861.1 GECZ01003194 JAS66575.1 CM000363 EDX09902.1 GEZM01068122 JAV67165.1 GFTR01004225 JAW12201.1 GBGD01002603 JAC86286.1 KMY98745.1 KQS43727.1 GDHF01018347 JAI33967.1 GAKP01007375 JAC51577.1 KQ979609 KYN20197.1 GL437252 EFN70765.1 KRK00845.1 KPU77885.1 GBXI01012097 JAD02195.1 JXJN01004319 KRG05950.1 LJIG01022892 KRT78193.1 GDHF01007958 JAI44356.1 KRF84386.1 DS231933 EDS27564.1 BT127367 AEE62329.1 GBXI01008554 JAD05738.1
OWR42197.1 KQ461190 KPJ07232.1 KQ459590 KPI97311.1 JTDY01004183 KOB68488.1 GEZM01068121 JAV67166.1 KQ971329 KYB28350.1 GDUN01000180 JAN95739.1 GFDL01000876 JAV34169.1 GEZM01068118 JAV67168.1 GGFM01004972 MBW25723.1 GGFJ01008636 MBW57777.1 GGFK01010998 MBW44319.1 GGFL01001597 MBW65775.1 GGFL01001595 MBW65773.1 GGFL01001596 MBW65774.1 GFDF01009940 JAV04144.1 JXUM01107731 KQ565169 KXJ71257.1 GAPW01004312 JAC09286.1 KK853239 KDR09509.1 GANO01001522 JAB58349.1 CM002912 KMY98744.1 AE014296 AY060437 BT001491 AAL25476.1 AAS65049.1 CH954178 EDV51247.1 CH480815 EDW40920.1 GDAI01002770 JAI14833.1 CM000159 EDW92967.1 GEZM01068119 JAV67167.1 EFA01153.1 GBHO01021469 GBRD01010193 GDHC01010432 GDHC01008823 GDHC01000313 JAG22135.1 JAG55631.1 JAQ08197.1 JAQ09806.1 JAQ18316.1 CH902618 EDV39088.1 CH933809 EDW18083.1 GL453568 EFN75873.1 AAAB01008900 CH940647 EDW69415.1 KQ414784 KOC61129.1 KQ982585 KYQ54326.1 GGFL01016299 MBW80477.1 CH916366 EDV96037.1 KQ981979 KYN31442.1 GGFM01007737 MBW28488.1 GL887491 EGI71095.1 KQ976625 KYM79058.1 CH964095 EDW78975.1 ADTU01001538 KQ976932 KYN07038.1 GL762556 EFZ21140.1 AXCM01001477 GEDC01031410 JAS05888.1 KK107847 QOIP01000005 EZA47485.1 RLU23041.1 CH379069 EDY73726.1 CH479240 EDW37554.1 KQ435711 KOX79479.1 KZ288210 PBC32923.1 JR046405 AEY60186.1 EAA09480.5 CAQQ02182124 CAQQ02182125 GEBQ01018162 JAT21815.1 APCN01002823 AJVK01026926 GECU01020845 JAS86861.1 GECZ01003194 JAS66575.1 CM000363 EDX09902.1 GEZM01068122 JAV67165.1 GFTR01004225 JAW12201.1 GBGD01002603 JAC86286.1 KMY98745.1 KQS43727.1 GDHF01018347 JAI33967.1 GAKP01007375 JAC51577.1 KQ979609 KYN20197.1 GL437252 EFN70765.1 KRK00845.1 KPU77885.1 GBXI01012097 JAD02195.1 JXJN01004319 KRG05950.1 LJIG01022892 KRT78193.1 GDHF01007958 JAI44356.1 KRF84386.1 DS231933 EDS27564.1 BT127367 AEE62329.1 GBXI01008554 JAD05738.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000037510
UP000007266
+ More
UP000075880 UP000076407 UP000075902 UP000069940 UP000249989 UP000027135 UP000000803 UP000008711 UP000001292 UP000002282 UP000007801 UP000192221 UP000009192 UP000075901 UP000008237 UP000008792 UP000095301 UP000053825 UP000075809 UP000001070 UP000078541 UP000075900 UP000007755 UP000078540 UP000007798 UP000005205 UP000078542 UP000075883 UP000075884 UP000053097 UP000279307 UP000001819 UP000008744 UP000095300 UP000053105 UP000242457 UP000005203 UP000007062 UP000002358 UP000015102 UP000075840 UP000092462 UP000000304 UP000078492 UP000000311 UP000092443 UP000092460 UP000002320
UP000075880 UP000076407 UP000075902 UP000069940 UP000249989 UP000027135 UP000000803 UP000008711 UP000001292 UP000002282 UP000007801 UP000192221 UP000009192 UP000075901 UP000008237 UP000008792 UP000095301 UP000053825 UP000075809 UP000001070 UP000078541 UP000075900 UP000007755 UP000078540 UP000007798 UP000005205 UP000078542 UP000075883 UP000075884 UP000053097 UP000279307 UP000001819 UP000008744 UP000095300 UP000053105 UP000242457 UP000005203 UP000007062 UP000002358 UP000015102 UP000075840 UP000092462 UP000000304 UP000078492 UP000000311 UP000092443 UP000092460 UP000002320
Pfam
PF00583 Acetyltransf_1
SUPFAM
SSF55729
SSF55729
ProteinModelPortal
H9JQ63
A0A2W1BRR9
A0A2H1VSZ3
A0A212EL51
A0A194QNZ0
A0A194PV85
+ More
A0A0L7KYZ1 A0A1Y1L6H4 A0A139WK49 A0A0P6J6A3 A0A1Q3G351 A0A182JG47 A0A182XEX5 A0A1Y1L305 A0A2M3ZB36 A0A2M4BXH6 A0A2M4AU45 A0A2M4CKK6 A0A2M4CKE3 A0A2M4CKC5 A0A1Y9INA8 A0A1L8DCV4 A0A182H3N9 A0A023EJQ4 A0A067QKR8 U5EL81 A0A0J9RTX3 Q95SX8-2 B3NGL1 B4HLA1 A0A0K8TKZ5 B4PEC9 A0A1Y1L7W5 D6WGD7 A0A0A9XY07 B3MA74 A0A1W4U9U8 B4L0K6 A0A182T5X9 E2C870 A0A1S4GND8 B4LGA7 A0A1I8NF75 A0A0L7QRE9 A0A151X239 A0A2M4DSE6 B4J2P6 A0A195EU61 A0A182RX08 A0A2M3ZJ68 F4W499 A0A195B3J0 B4N4A1 A0A158NFW6 A0A151INW9 E9IE22 A0A182M1M3 A0A182N1Q6 A0A1B6BXA0 A0A026VUY1 B5DQ59 B4HAF6 A0A1I8QAL0 A0A0M9AB05 A0A2A3EM93 V9II72 A0A088AAN9 Q7PQ00 K7IR38 T1GN36 A0A1B6LDQ0 A0A182HP01 A0A1B0D7I0 A0A1B6IIX5 A0A1B6GW07 B4QNN5 A0A1Y1L5A3 Q95SX8 A0A224XW67 A0A069DRG7 A0A0J9UIB6 A0A0Q5U3R3 A0A0K8V4V0 A0A034WBA5 A0A195E5M6 E2A733 A0A0R1DUX1 Q95SX8-3 A0A0P8YG23 A0A0A1WUK0 A0A1W4UL81 A0A1A9YEB9 A0A1B0AVM4 A0A0Q9XLY8 A0A0T6ASX3 A0A0K8VZU5 A0A0Q9WJ68 B0WH57 J3JVU9 A0A0A1X4X6
A0A0L7KYZ1 A0A1Y1L6H4 A0A139WK49 A0A0P6J6A3 A0A1Q3G351 A0A182JG47 A0A182XEX5 A0A1Y1L305 A0A2M3ZB36 A0A2M4BXH6 A0A2M4AU45 A0A2M4CKK6 A0A2M4CKE3 A0A2M4CKC5 A0A1Y9INA8 A0A1L8DCV4 A0A182H3N9 A0A023EJQ4 A0A067QKR8 U5EL81 A0A0J9RTX3 Q95SX8-2 B3NGL1 B4HLA1 A0A0K8TKZ5 B4PEC9 A0A1Y1L7W5 D6WGD7 A0A0A9XY07 B3MA74 A0A1W4U9U8 B4L0K6 A0A182T5X9 E2C870 A0A1S4GND8 B4LGA7 A0A1I8NF75 A0A0L7QRE9 A0A151X239 A0A2M4DSE6 B4J2P6 A0A195EU61 A0A182RX08 A0A2M3ZJ68 F4W499 A0A195B3J0 B4N4A1 A0A158NFW6 A0A151INW9 E9IE22 A0A182M1M3 A0A182N1Q6 A0A1B6BXA0 A0A026VUY1 B5DQ59 B4HAF6 A0A1I8QAL0 A0A0M9AB05 A0A2A3EM93 V9II72 A0A088AAN9 Q7PQ00 K7IR38 T1GN36 A0A1B6LDQ0 A0A182HP01 A0A1B0D7I0 A0A1B6IIX5 A0A1B6GW07 B4QNN5 A0A1Y1L5A3 Q95SX8 A0A224XW67 A0A069DRG7 A0A0J9UIB6 A0A0Q5U3R3 A0A0K8V4V0 A0A034WBA5 A0A195E5M6 E2A733 A0A0R1DUX1 Q95SX8-3 A0A0P8YG23 A0A0A1WUK0 A0A1W4UL81 A0A1A9YEB9 A0A1B0AVM4 A0A0Q9XLY8 A0A0T6ASX3 A0A0K8VZU5 A0A0Q9WJ68 B0WH57 J3JVU9 A0A0A1X4X6
PDB
5HH0
E-value=3.17029e-59,
Score=577
Ontologies
KEGG
GO
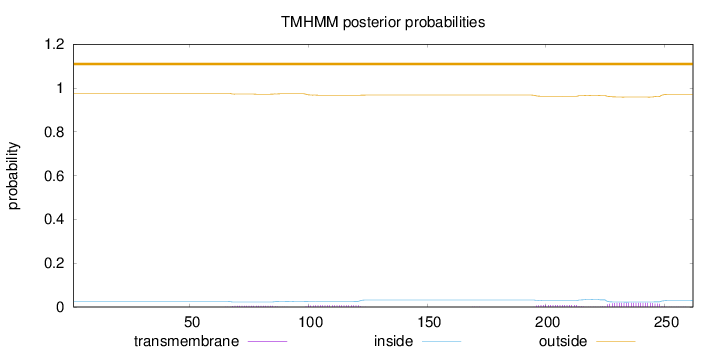
Topology
Length:
262
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.87222
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02461
outside
1 - 262
Population Genetic Test Statistics
Pi
284.832592
Theta
203.286907
Tajima's D
0.143818
CLR
0.203568
CSRT
0.411679416029199
Interpretation
Uncertain
