Gene
KWMTBOMO06583
Pre Gene Modal
BGIBMGA011808
Annotation
PREDICTED:_lariat_debranching_enzyme_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.925
Sequence
CDS
ATGAAAATAGCAGTGGAAGGATGCGCTCATGGAGAATTAGACAAAATCTACGATTGCATAGAAACATTACAAGAGAGAGAAGGTTTTAAGATTGATTTGTTAATATGTTGCGGTGACTTTCAATCTGTACGAAATGCCGCTGATTTACGTGCAATGGCTGTACCCGAGAAATATCAACGCATGTGCACATTTTACAAATATTACAGTGGGGAGAAGAAGGCACCTGTATTAACATTATTTATTGGTGGAAACCATGAAGCTTCAAACTATCTACAGGAGCTTCCGTATGGTGGGTGGGTGGCTCCCAATATATACTACATGGGACGTGCTGGTGTTGTTAAGTTTGGAGAACTGAGGATTGGTGGATTATCAGGAATATTCAAAGGGCATGATTACCTGCAAGGCCTCTGGGAATGTTTGCCCTACAACCAAAACTCCATGAGGTCTGTGTATCATATCAGGTCACTAGATGTCTTTAGATTATCTCAATTGAAAGAAAAACTACATGTAATGTTGACACATGACTGGCCTCGTGGTATTACAGATTATGGAGACAAAGATCACTTGTTGAGAAGAAAACCTTTTTTAAGGGAAGATATTGAATCAAACCAATTAGGTAGTATGCCTGCAGAAAAATTACTTCATACATTGAAACCAGATTACTGGTTTGCAGCACATTTGCACTGTCAATTTGCAGCATTGGTCAAACATGAAGATAACAGGGAAACGAAGTTTCTTGCTCTGGACAAATGTTTGCCAAAACGTAGACATTTACAAATATTAGATCTACCTCATGAGTATGATGGAGATAAATGCTTGAAACATGATGCAGAATGGTTAGTTGTGTTGAAGAATACAAATCATCTGCTGAATGTTAAAAACATGGAGGGTCATCTACCTGGACCGGGCGGAAATGAAAGGTACAATTTTACCCCAACCAATGAAGAAAAAGAAATTTTAATTCAGTCCTTGCAAAATTTGGCTATTACATCTGAAAATTTCGTCAAAACTGCACCTGCATACAAGCCAGGAGCTCCCAAAGCATATCTCACAGACCCTATACTAAATCCACAGACAGTAGATTTATGTGAGAAATTAGGTATTGATGATCCTGTACAAGTGATTATGGCACGAGCAGGCAGAGTTATGGAGAAACCAACTGATGTCGATATTGTTGAAACGAAAGCAGATCTCAATCCAGTACTAAAAACACCTATGTCAAAACTAATTTTACCAGCACCTGTGACACCTAGTGAAAATGATTGTGAAGAAAATACATGTACAACACCTATTTCAGAAAATCCCTTATTATTAGATAGTTTATGTAATACTTCTGAATGTACGACTCCTACTTCAATTAGTGGGAAGAAAACATTCAAGAGGCGAAATATATCAATTTATAGTTCCACAGAAGAAGAGAGTCAAAGTATGAACTCAAGTTCCTTAGTAAGTCCAACCAGTCCATCAAAATATCCTAGGGGTAATATTGAATAA
Protein
MKIAVEGCAHGELDKIYDCIETLQEREGFKIDLLICCGDFQSVRNAADLRAMAVPEKYQRMCTFYKYYSGEKKAPVLTLFIGGNHEASNYLQELPYGGWVAPNIYYMGRAGVVKFGELRIGGLSGIFKGHDYLQGLWECLPYNQNSMRSVYHIRSLDVFRLSQLKEKLHVMLTHDWPRGITDYGDKDHLLRRKPFLREDIESNQLGSMPAEKLLHTLKPDYWFAAHLHCQFAALVKHEDNRETKFLALDKCLPKRRHLQILDLPHEYDGDKCLKHDAEWLVVLKNTNHLLNVKNMEGHLPGPGGNERYNFTPTNEEKEILIQSLQNLAITSENFVKTAPAYKPGAPKAYLTDPILNPQTVDLCEKLGIDDPVQVIMARAGRVMEKPTDVDIVETKADLNPVLKTPMSKLILPAPVTPSENDCEENTCTTPISENPLLLDSLCNTSECTTPTSISGKKTFKRRNISIYSSTEEESQSMNSSSLVSPTSPSKYPRGNIE
Summary
Uniprot
H9JQK2
A0A3S2LUF9
S4NSN7
A0A2A4J4Y8
A0A1E1WKW8
A0A212EL36
+ More
A0A194QQI3 A0A0L7L1U3 A0A194PWD3 A0A2H1VG55 A0A2W1BLA8 A0A2W1BP07 A0A1Q3EYY6 U4UA57 N6U4P2 B0WC77 A0A182GF62 D6WDD9 A0A336LVS1 W5JGV9 A0A0P6JRZ8 A0A3F2YQ97 A0A336MVD9 A0A1B6LZV8 A0A1L8DFX6 A0A069DVC4 A0A182K0Q0 A0A182MJ64 A0A182PIW0 A0A0P5TF60 A0A182WI35 A0A0P5FXV0 A0A0P5VUY7 A0A164W622 A0A1B0C9U2 A0A0V0GBC9 A0A1B6DWG1 A0A182R6P7 A0A0P5C9K7 A0A0P5XMW2 A0A1B6CUA3 A0A1W4XE65 A0A1B6FR46 A0A0L0C1Y1 A0A182Y888 A0A182N7F9 A0A1Z5KXJ4 E9HAB2 A0A2J7PPH4 A0A2J7PPC9 A0A084WMW3 A0A182QEL4 A0A1I8Q5G1 A0A182I6D5 A0A182L402 A0A210QH95 A0A182X275 A0A182VNZ8 A0A182U883 A0A182IY73 A0A195F1D7 A0A0P5TKD7 Q7QIE4 E2A9L0 A0A026WSM6 A0A0P5UZJ4 E0VAQ8 A0A1I8M588 A0A2R5LB18 A0A0P5PPK5 A0A151XB78 K1Q687 A0A2S2QUT4 A0A067RBG6 W8BB13 A0A0P5XEW6 A0A293LZ91 A0A0P4WD94 A0A151I1B3 A0A0N7Z9E9 A0A0P5WT81 A0A0P5BHJ6 A0A195E6Z3 A0A0P6BCB4 J9JWY2 A0A0J7NEI1 A0A2A3EAV4 T1I1S4 A0A3R7N5D6 A0A088AHU7 A0A0P5XBA3 A0A1Y1N083 A0A0P5QL06 V5HSG9 A0A0L7RIE9 A0A131Y1N5 B3M423 A0A0N0U6G0 A0A0A1XSH9 F4X6K4
A0A194QQI3 A0A0L7L1U3 A0A194PWD3 A0A2H1VG55 A0A2W1BLA8 A0A2W1BP07 A0A1Q3EYY6 U4UA57 N6U4P2 B0WC77 A0A182GF62 D6WDD9 A0A336LVS1 W5JGV9 A0A0P6JRZ8 A0A3F2YQ97 A0A336MVD9 A0A1B6LZV8 A0A1L8DFX6 A0A069DVC4 A0A182K0Q0 A0A182MJ64 A0A182PIW0 A0A0P5TF60 A0A182WI35 A0A0P5FXV0 A0A0P5VUY7 A0A164W622 A0A1B0C9U2 A0A0V0GBC9 A0A1B6DWG1 A0A182R6P7 A0A0P5C9K7 A0A0P5XMW2 A0A1B6CUA3 A0A1W4XE65 A0A1B6FR46 A0A0L0C1Y1 A0A182Y888 A0A182N7F9 A0A1Z5KXJ4 E9HAB2 A0A2J7PPH4 A0A2J7PPC9 A0A084WMW3 A0A182QEL4 A0A1I8Q5G1 A0A182I6D5 A0A182L402 A0A210QH95 A0A182X275 A0A182VNZ8 A0A182U883 A0A182IY73 A0A195F1D7 A0A0P5TKD7 Q7QIE4 E2A9L0 A0A026WSM6 A0A0P5UZJ4 E0VAQ8 A0A1I8M588 A0A2R5LB18 A0A0P5PPK5 A0A151XB78 K1Q687 A0A2S2QUT4 A0A067RBG6 W8BB13 A0A0P5XEW6 A0A293LZ91 A0A0P4WD94 A0A151I1B3 A0A0N7Z9E9 A0A0P5WT81 A0A0P5BHJ6 A0A195E6Z3 A0A0P6BCB4 J9JWY2 A0A0J7NEI1 A0A2A3EAV4 T1I1S4 A0A3R7N5D6 A0A088AHU7 A0A0P5XBA3 A0A1Y1N083 A0A0P5QL06 V5HSG9 A0A0L7RIE9 A0A131Y1N5 B3M423 A0A0N0U6G0 A0A0A1XSH9 F4X6K4
Pubmed
19121390
23622113
22118469
26354079
26227816
28756777
+ More
23537049 26483478 18362917 19820115 20920257 23761445 26999592 26334808 26108605 25244985 28528879 21292972 24438588 20966253 28812685 12364791 14747013 17210077 20798317 24508170 20566863 25315136 22992520 24845553 24495485 27129103 28004739 25765539 17994087 25830018 21719571
23537049 26483478 18362917 19820115 20920257 23761445 26999592 26334808 26108605 25244985 28528879 21292972 24438588 20966253 28812685 12364791 14747013 17210077 20798317 24508170 20566863 25315136 22992520 24845553 24495485 27129103 28004739 25765539 17994087 25830018 21719571
EMBL
BABH01012081
RSAL01000209
RVE44245.1
GAIX01013997
JAA78563.1
NWSH01003314
+ More
PCG66574.1 GDQN01003384 JAT87670.1 AGBW02014142 OWR42195.1 KQ461190 KPJ07230.1 JTDY01003602 KOB69281.1 KQ459590 KPI97313.1 ODYU01002361 SOQ39761.1 KZ150049 PZC74455.1 KZ149973 PZC76001.1 GFDL01014531 JAV20514.1 KB631957 ERL87471.1 APGK01043139 KB741011 ENN75576.1 DS231886 EDS43322.1 JXUM01059292 KQ562045 KXJ76820.1 KQ971321 EEZ99472.1 UFQT01000224 SSX22035.1 ADMH02001223 ETN63597.1 GDUN01000824 JAN95095.1 UFQS01002111 UFQT01002111 SSX13265.1 SSX32703.1 GEBQ01013282 GEBQ01010807 JAT26695.1 JAT29170.1 GFDF01008726 JAV05358.1 GBGD01001123 JAC87766.1 AXCM01002040 GDIP01130850 JAL72864.1 GDIQ01249894 JAK01831.1 GDIP01095946 JAM07769.1 LRGB01001348 KZS12981.1 AJWK01002798 GECL01000883 JAP05241.1 GEDC01007292 JAS30006.1 GDIP01189838 JAJ33564.1 GDIP01082093 JAM21622.1 GEDC01020485 JAS16813.1 GECZ01017101 JAS52668.1 JRES01000997 KNC26266.1 GFJQ02007459 JAV99510.1 GL732611 EFX71275.1 NEVH01022715 PNF18199.1 PNF18204.1 ATLV01024521 KE525352 KFB51557.1 AXCN02000573 APCN01003620 NEDP02003668 OWF48133.1 KQ981864 KYN34191.1 GDIP01125096 JAL78618.1 AAAB01008807 EAA04183.4 GL437917 EFN69900.1 KK107109 EZA59050.1 GDIP01107153 JAL96561.1 DS235012 EEB10464.1 GGLE01002532 MBY06658.1 GDIQ01125734 JAL25992.1 KQ982335 KYQ57607.1 JH818630 EKC26794.1 GGMS01012313 MBY81516.1 KK852567 KDR21191.1 GAMC01016214 JAB90341.1 GDIP01074689 JAM29026.1 GFWV01008647 MAA33376.1 GDRN01067002 JAI64442.1 KQ976580 KYM79900.1 GDKW01000739 JAI55856.1 GDIP01082092 JAM21623.1 GDIP01189837 JAJ33565.1 KQ979568 KYN20958.1 GDIP01017267 JAM86448.1 ABLF02033601 LBMM01006104 KMQ90935.1 KZ288310 PBC28604.1 ACPB03014728 QCYY01001486 ROT77735.1 GDIP01076188 JAM27527.1 GEZM01018711 JAV90045.1 GDIQ01119136 JAL32590.1 GANP01003214 JAB81254.1 KQ414583 KOC70742.1 GEFM01003411 JAP72385.1 CH902618 EDV40385.1 KQ435727 KOX77928.1 GBXI01008331 GBXI01000316 JAD05961.1 JAD13976.1 GL888818 EGI57964.1
PCG66574.1 GDQN01003384 JAT87670.1 AGBW02014142 OWR42195.1 KQ461190 KPJ07230.1 JTDY01003602 KOB69281.1 KQ459590 KPI97313.1 ODYU01002361 SOQ39761.1 KZ150049 PZC74455.1 KZ149973 PZC76001.1 GFDL01014531 JAV20514.1 KB631957 ERL87471.1 APGK01043139 KB741011 ENN75576.1 DS231886 EDS43322.1 JXUM01059292 KQ562045 KXJ76820.1 KQ971321 EEZ99472.1 UFQT01000224 SSX22035.1 ADMH02001223 ETN63597.1 GDUN01000824 JAN95095.1 UFQS01002111 UFQT01002111 SSX13265.1 SSX32703.1 GEBQ01013282 GEBQ01010807 JAT26695.1 JAT29170.1 GFDF01008726 JAV05358.1 GBGD01001123 JAC87766.1 AXCM01002040 GDIP01130850 JAL72864.1 GDIQ01249894 JAK01831.1 GDIP01095946 JAM07769.1 LRGB01001348 KZS12981.1 AJWK01002798 GECL01000883 JAP05241.1 GEDC01007292 JAS30006.1 GDIP01189838 JAJ33564.1 GDIP01082093 JAM21622.1 GEDC01020485 JAS16813.1 GECZ01017101 JAS52668.1 JRES01000997 KNC26266.1 GFJQ02007459 JAV99510.1 GL732611 EFX71275.1 NEVH01022715 PNF18199.1 PNF18204.1 ATLV01024521 KE525352 KFB51557.1 AXCN02000573 APCN01003620 NEDP02003668 OWF48133.1 KQ981864 KYN34191.1 GDIP01125096 JAL78618.1 AAAB01008807 EAA04183.4 GL437917 EFN69900.1 KK107109 EZA59050.1 GDIP01107153 JAL96561.1 DS235012 EEB10464.1 GGLE01002532 MBY06658.1 GDIQ01125734 JAL25992.1 KQ982335 KYQ57607.1 JH818630 EKC26794.1 GGMS01012313 MBY81516.1 KK852567 KDR21191.1 GAMC01016214 JAB90341.1 GDIP01074689 JAM29026.1 GFWV01008647 MAA33376.1 GDRN01067002 JAI64442.1 KQ976580 KYM79900.1 GDKW01000739 JAI55856.1 GDIP01082092 JAM21623.1 GDIP01189837 JAJ33565.1 KQ979568 KYN20958.1 GDIP01017267 JAM86448.1 ABLF02033601 LBMM01006104 KMQ90935.1 KZ288310 PBC28604.1 ACPB03014728 QCYY01001486 ROT77735.1 GDIP01076188 JAM27527.1 GEZM01018711 JAV90045.1 GDIQ01119136 JAL32590.1 GANP01003214 JAB81254.1 KQ414583 KOC70742.1 GEFM01003411 JAP72385.1 CH902618 EDV40385.1 KQ435727 KOX77928.1 GBXI01008331 GBXI01000316 JAD05961.1 JAD13976.1 GL888818 EGI57964.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000037510
+ More
UP000053268 UP000030742 UP000019118 UP000002320 UP000069940 UP000249989 UP000007266 UP000000673 UP000069272 UP000075881 UP000075883 UP000075885 UP000075920 UP000076858 UP000092461 UP000075900 UP000192223 UP000037069 UP000076408 UP000075884 UP000000305 UP000235965 UP000030765 UP000075886 UP000095300 UP000075840 UP000075882 UP000242188 UP000076407 UP000075903 UP000075902 UP000075880 UP000078541 UP000007062 UP000000311 UP000053097 UP000009046 UP000095301 UP000075809 UP000005408 UP000027135 UP000078540 UP000078492 UP000007819 UP000036403 UP000242457 UP000015103 UP000283509 UP000005203 UP000053825 UP000007801 UP000053105 UP000007755
UP000053268 UP000030742 UP000019118 UP000002320 UP000069940 UP000249989 UP000007266 UP000000673 UP000069272 UP000075881 UP000075883 UP000075885 UP000075920 UP000076858 UP000092461 UP000075900 UP000192223 UP000037069 UP000076408 UP000075884 UP000000305 UP000235965 UP000030765 UP000075886 UP000095300 UP000075840 UP000075882 UP000242188 UP000076407 UP000075903 UP000075902 UP000075880 UP000078541 UP000007062 UP000000311 UP000053097 UP000009046 UP000095301 UP000075809 UP000005408 UP000027135 UP000078540 UP000078492 UP000007819 UP000036403 UP000242457 UP000015103 UP000283509 UP000005203 UP000053825 UP000007801 UP000053105 UP000007755
PRIDE
Pfam
Interpro
IPR041816
Dbr1_N
+ More
IPR004843 Calcineurin-like_PHP_ApaH
IPR029052 Metallo-depent_PP-like
IPR007708 DBR1_C
IPR006203 GHMP_knse_ATP-bd_CS
IPR006204 GHMP_kinase_N_dom
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR019741 Galactokinase_CS
IPR019539 GalKase_gal-bd
IPR000705 Galactokinase
IPR020568 Ribosomal_S5_D2-typ_fold
IPR037663 Mosmo
IPR004843 Calcineurin-like_PHP_ApaH
IPR029052 Metallo-depent_PP-like
IPR007708 DBR1_C
IPR006203 GHMP_knse_ATP-bd_CS
IPR006204 GHMP_kinase_N_dom
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR019741 Galactokinase_CS
IPR019539 GalKase_gal-bd
IPR000705 Galactokinase
IPR020568 Ribosomal_S5_D2-typ_fold
IPR037663 Mosmo
SUPFAM
SSF54211
SSF54211
Gene 3D
ProteinModelPortal
H9JQK2
A0A3S2LUF9
S4NSN7
A0A2A4J4Y8
A0A1E1WKW8
A0A212EL36
+ More
A0A194QQI3 A0A0L7L1U3 A0A194PWD3 A0A2H1VG55 A0A2W1BLA8 A0A2W1BP07 A0A1Q3EYY6 U4UA57 N6U4P2 B0WC77 A0A182GF62 D6WDD9 A0A336LVS1 W5JGV9 A0A0P6JRZ8 A0A3F2YQ97 A0A336MVD9 A0A1B6LZV8 A0A1L8DFX6 A0A069DVC4 A0A182K0Q0 A0A182MJ64 A0A182PIW0 A0A0P5TF60 A0A182WI35 A0A0P5FXV0 A0A0P5VUY7 A0A164W622 A0A1B0C9U2 A0A0V0GBC9 A0A1B6DWG1 A0A182R6P7 A0A0P5C9K7 A0A0P5XMW2 A0A1B6CUA3 A0A1W4XE65 A0A1B6FR46 A0A0L0C1Y1 A0A182Y888 A0A182N7F9 A0A1Z5KXJ4 E9HAB2 A0A2J7PPH4 A0A2J7PPC9 A0A084WMW3 A0A182QEL4 A0A1I8Q5G1 A0A182I6D5 A0A182L402 A0A210QH95 A0A182X275 A0A182VNZ8 A0A182U883 A0A182IY73 A0A195F1D7 A0A0P5TKD7 Q7QIE4 E2A9L0 A0A026WSM6 A0A0P5UZJ4 E0VAQ8 A0A1I8M588 A0A2R5LB18 A0A0P5PPK5 A0A151XB78 K1Q687 A0A2S2QUT4 A0A067RBG6 W8BB13 A0A0P5XEW6 A0A293LZ91 A0A0P4WD94 A0A151I1B3 A0A0N7Z9E9 A0A0P5WT81 A0A0P5BHJ6 A0A195E6Z3 A0A0P6BCB4 J9JWY2 A0A0J7NEI1 A0A2A3EAV4 T1I1S4 A0A3R7N5D6 A0A088AHU7 A0A0P5XBA3 A0A1Y1N083 A0A0P5QL06 V5HSG9 A0A0L7RIE9 A0A131Y1N5 B3M423 A0A0N0U6G0 A0A0A1XSH9 F4X6K4
A0A194QQI3 A0A0L7L1U3 A0A194PWD3 A0A2H1VG55 A0A2W1BLA8 A0A2W1BP07 A0A1Q3EYY6 U4UA57 N6U4P2 B0WC77 A0A182GF62 D6WDD9 A0A336LVS1 W5JGV9 A0A0P6JRZ8 A0A3F2YQ97 A0A336MVD9 A0A1B6LZV8 A0A1L8DFX6 A0A069DVC4 A0A182K0Q0 A0A182MJ64 A0A182PIW0 A0A0P5TF60 A0A182WI35 A0A0P5FXV0 A0A0P5VUY7 A0A164W622 A0A1B0C9U2 A0A0V0GBC9 A0A1B6DWG1 A0A182R6P7 A0A0P5C9K7 A0A0P5XMW2 A0A1B6CUA3 A0A1W4XE65 A0A1B6FR46 A0A0L0C1Y1 A0A182Y888 A0A182N7F9 A0A1Z5KXJ4 E9HAB2 A0A2J7PPH4 A0A2J7PPC9 A0A084WMW3 A0A182QEL4 A0A1I8Q5G1 A0A182I6D5 A0A182L402 A0A210QH95 A0A182X275 A0A182VNZ8 A0A182U883 A0A182IY73 A0A195F1D7 A0A0P5TKD7 Q7QIE4 E2A9L0 A0A026WSM6 A0A0P5UZJ4 E0VAQ8 A0A1I8M588 A0A2R5LB18 A0A0P5PPK5 A0A151XB78 K1Q687 A0A2S2QUT4 A0A067RBG6 W8BB13 A0A0P5XEW6 A0A293LZ91 A0A0P4WD94 A0A151I1B3 A0A0N7Z9E9 A0A0P5WT81 A0A0P5BHJ6 A0A195E6Z3 A0A0P6BCB4 J9JWY2 A0A0J7NEI1 A0A2A3EAV4 T1I1S4 A0A3R7N5D6 A0A088AHU7 A0A0P5XBA3 A0A1Y1N083 A0A0P5QL06 V5HSG9 A0A0L7RIE9 A0A131Y1N5 B3M423 A0A0N0U6G0 A0A0A1XSH9 F4X6K4
PDB
5UKI
E-value=1.8518e-49,
Score=496
Ontologies
GO
Topology
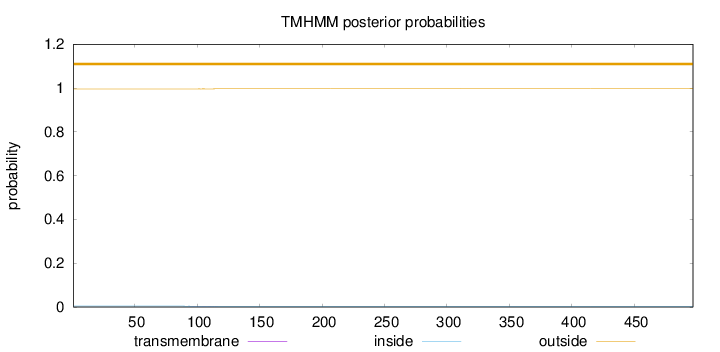
Length:
497
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01365
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00406
outside
1 - 497
Population Genetic Test Statistics
Pi
179.145422
Theta
169.673914
Tajima's D
0.47567
CLR
0.375409
CSRT
0.508624568771561
Interpretation
Uncertain
