Gene
KWMTBOMO06581
Pre Gene Modal
BGIBMGA011666
Annotation
PREDICTED:_uncharacterized_protein_LOC105842372_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.149 Nuclear Reliability : 1.546
Sequence
CDS
ATGTGTAGTCATCCCCACGGTGCCCGTGTGTCCGGCGTGGTCGAGGAGACGCCAGGACGTCTTGAATACGAAGTAGCTCTGTACAAGGAAGCACCGCCTGTCACAAGACATTCTAATCATTCGGTTGATTTGCCTGTCGATCAGGCTGTTCCAATCGGCACGAAGCTTCAACTACGCGCACGGATTAATCCCGACTCAGCGTGGCGGCACATCAAACTGCTGGAGGTAGCCGTGTCTCCTGACCCTGACAGACCTCACGCACCCGGATCCGTTTTGCTCGTTAAAGACGGATGCCGCAATCGGGATTTCGCTTCGATAATCCCGCATCAACCAGCGCGCTACCGGGAGCGACATAACGAAGTATTCTTGGACTTTGAAGCTTTCCTGCTGGCTTCTATGAAGGAGCGCTCAACGTTATGGATACACTCGCAGATAAAAGCCTGCATGGACGCCGCAGACTGCCAACCAGACTACTGCCTCGACCTATTTGAACCATCAGGCCACGGAAGAAAAAGAAGGTCACTGCCGGAAACGCATAACGCTACACACGCAGCGCTAGTCAACGACAACTCGACAACGCCCTTCACGAGATTCAAGGAGAATCTAGAATACACAGTGGTCATGCCCGGAGAACTCTTTCATAGGACCCCTGTTGAAGCCACCTGTGCCACTTCGATGATGGTCGCCGTAGCACTCGGTGCACTGCTCTTCATGTCTGCATTACTAATGTGTTACTTAGCCACAAAATTAAATTCTACTTTATTGAAGAACAACAACCTTCAATCGCCAACCGGTAAAGGATTTGAACAGATTTTAAGGGAGCTGGCACACCATTCCTTACCGGATACGGGTTATACAGGACGGCCTACTGTACAATAG
Protein
MCSHPHGARVSGVVEETPGRLEYEVALYKEAPPVTRHSNHSVDLPVDQAVPIGTKLQLRARINPDSAWRHIKLLEVAVSPDPDRPHAPGSVLLVKDGCRNRDFASIIPHQPARYRERHNEVFLDFEAFLLASMKERSTLWIHSQIKACMDAADCQPDYCLDLFEPSGHGRKRRSLPETHNATHAALVNDNSTTPFTRFKENLEYTVVMPGELFHRTPVEATCATSMMVAVALGALLFMSALLMCYLATKLNSTLLKNNNLQSPTGKGFEQILRELAHHSLPDTGYTGRPTVQ
Summary
Uniprot
H9JQ60
A0A2A4J357
A0A2H1VFX9
A0A2A4J4B7
A0A2W1BHX7
A0A194QNX7
+ More
A0A194Q1N8 A0A212F7Y6 A0A0L7L1A7 D6WG56 A0A310SBM7 A0A2J7QAT2 A0A0L7QR14 A0A0N0U4I9 A0A087ZTV2 A0A2A3EGC3 A0A026X2F1 A0A2S2Q4W8 A0A3L8DSN7 E2C9Q5 A0A158P1F5 A0A195D127 A0A151WZB9 F4WYK3 A0A195E2S8 A0A195F3C3 E0VAT5 A0A151I2G3 A0A1W4WYZ4 E2AQE0 K7J869 T1HGT8 N6TWJ7 J9JQF1 A0A2P8XK82 A0A067R3Y2 U4U9V9 A0A1B6C7F8 A0A1B0CYH6 A0A1B0CRG5 A0A182SIP2 A0A182UK06 A0A182I7M5 A0A1S4H0N1 A0A2R7VZB5 A0A182VNS6 A0A182LME4 A0A182YFS5 A0A232EIB4 A0A182QRG8 A0A336L1F0 A0A182NBF0 A0A182PUW5 A0A182WXR2 A0A182KDD8 A0A182JJX8 A0A182VU18 A0A336L5G1 A0A084VKI4 A0A182RLC1 A0A1S4FYD2 Q16K05 A0A182H6D4 A0A182FNC9 A0A0J7NGC7 A0A182LYJ8 A0A2M3ZAA6 A0A1J1IKX8 W5J4U7 A0A1A9UTA7 B0XKL0 B0WXH2 A0A1B0AV56 A0A1B0FPX0 A0A2M3ZAV4 A0A1A9Y5Q7 A7UV83 A0A1A9WJ38 B4KHT4 A0A0Q9WJE5 A0A1A9ZM09 B4JPY0 A0A0K2THS8 A0A0P5F8U7
A0A194Q1N8 A0A212F7Y6 A0A0L7L1A7 D6WG56 A0A310SBM7 A0A2J7QAT2 A0A0L7QR14 A0A0N0U4I9 A0A087ZTV2 A0A2A3EGC3 A0A026X2F1 A0A2S2Q4W8 A0A3L8DSN7 E2C9Q5 A0A158P1F5 A0A195D127 A0A151WZB9 F4WYK3 A0A195E2S8 A0A195F3C3 E0VAT5 A0A151I2G3 A0A1W4WYZ4 E2AQE0 K7J869 T1HGT8 N6TWJ7 J9JQF1 A0A2P8XK82 A0A067R3Y2 U4U9V9 A0A1B6C7F8 A0A1B0CYH6 A0A1B0CRG5 A0A182SIP2 A0A182UK06 A0A182I7M5 A0A1S4H0N1 A0A2R7VZB5 A0A182VNS6 A0A182LME4 A0A182YFS5 A0A232EIB4 A0A182QRG8 A0A336L1F0 A0A182NBF0 A0A182PUW5 A0A182WXR2 A0A182KDD8 A0A182JJX8 A0A182VU18 A0A336L5G1 A0A084VKI4 A0A182RLC1 A0A1S4FYD2 Q16K05 A0A182H6D4 A0A182FNC9 A0A0J7NGC7 A0A182LYJ8 A0A2M3ZAA6 A0A1J1IKX8 W5J4U7 A0A1A9UTA7 B0XKL0 B0WXH2 A0A1B0AV56 A0A1B0FPX0 A0A2M3ZAV4 A0A1A9Y5Q7 A7UV83 A0A1A9WJ38 B4KHT4 A0A0Q9WJE5 A0A1A9ZM09 B4JPY0 A0A0K2THS8 A0A0P5F8U7
Pubmed
EMBL
BABH01012083
BABH01012084
NWSH01003314
PCG66577.1
ODYU01002361
SOQ39763.1
+ More
PCG66576.1 KZ150049 PZC74458.1 KQ461190 KPJ07228.1 KQ459590 KPI97315.1 AGBW02009815 OWR49852.1 JTDY01003602 KOB69283.1 KQ971320 EFA00531.2 KQ761729 OAD57157.1 NEVH01016327 PNF25693.1 KQ414784 KOC61070.1 KQ435831 KOX71701.1 KZ288268 PBC30051.1 KK107024 EZA62248.1 GGMS01003575 MBY72778.1 QOIP01000004 RLU23437.1 GL453897 EFN75295.1 ADTU01006521 ADTU01006522 KQ976986 KYN06608.1 KQ982642 KYQ53242.1 GL888450 EGI60735.1 KQ979709 KYN19453.1 KQ981852 KYN34963.1 DS235015 EEB10491.1 KQ976538 KYM81327.1 GL441723 EFN64354.1 ACPB03006856 APGK01049772 KB741156 ENN73640.1 ABLF02034737 ABLF02034739 PYGN01001869 PSN32418.1 KK852999 KDR12652.1 KB631941 ERL87356.1 GEDC01027944 JAS09354.1 AJVK01000241 AJVK01000242 AJVK01000243 AJVK01000244 AJWK01024781 AJWK01024782 APCN01003427 AAAB01008984 KK854188 PTY12827.1 NNAY01004299 OXU18085.1 AXCN02000624 UFQS01001299 UFQT01001299 SSX10170.1 SSX29891.1 UFQS01002083 UFQT01002083 SSX13166.1 SSX32605.1 ATLV01014225 KE524948 KFB38478.1 CH477982 EAT34620.1 JXUM01113965 KQ565740 KXJ70721.1 LBMM01005346 KMQ91590.1 AXCM01005820 GGFM01004690 MBW25441.1 CVRI01000054 CRL00895.1 ADMH02002194 ETN57885.1 DS233879 EDS32061.1 DS232164 EDS36502.1 JXJN01003997 JXJN01003998 CCAG010023386 GGFM01004913 MBW25664.1 EDO63384.1 CH933807 EDW13371.2 CH940649 KRF81038.1 CH916372 EDV98960.1 HACA01008128 CDW25489.1 GDIQ01258872 JAJ92852.1
PCG66576.1 KZ150049 PZC74458.1 KQ461190 KPJ07228.1 KQ459590 KPI97315.1 AGBW02009815 OWR49852.1 JTDY01003602 KOB69283.1 KQ971320 EFA00531.2 KQ761729 OAD57157.1 NEVH01016327 PNF25693.1 KQ414784 KOC61070.1 KQ435831 KOX71701.1 KZ288268 PBC30051.1 KK107024 EZA62248.1 GGMS01003575 MBY72778.1 QOIP01000004 RLU23437.1 GL453897 EFN75295.1 ADTU01006521 ADTU01006522 KQ976986 KYN06608.1 KQ982642 KYQ53242.1 GL888450 EGI60735.1 KQ979709 KYN19453.1 KQ981852 KYN34963.1 DS235015 EEB10491.1 KQ976538 KYM81327.1 GL441723 EFN64354.1 ACPB03006856 APGK01049772 KB741156 ENN73640.1 ABLF02034737 ABLF02034739 PYGN01001869 PSN32418.1 KK852999 KDR12652.1 KB631941 ERL87356.1 GEDC01027944 JAS09354.1 AJVK01000241 AJVK01000242 AJVK01000243 AJVK01000244 AJWK01024781 AJWK01024782 APCN01003427 AAAB01008984 KK854188 PTY12827.1 NNAY01004299 OXU18085.1 AXCN02000624 UFQS01001299 UFQT01001299 SSX10170.1 SSX29891.1 UFQS01002083 UFQT01002083 SSX13166.1 SSX32605.1 ATLV01014225 KE524948 KFB38478.1 CH477982 EAT34620.1 JXUM01113965 KQ565740 KXJ70721.1 LBMM01005346 KMQ91590.1 AXCM01005820 GGFM01004690 MBW25441.1 CVRI01000054 CRL00895.1 ADMH02002194 ETN57885.1 DS233879 EDS32061.1 DS232164 EDS36502.1 JXJN01003997 JXJN01003998 CCAG010023386 GGFM01004913 MBW25664.1 EDO63384.1 CH933807 EDW13371.2 CH940649 KRF81038.1 CH916372 EDV98960.1 HACA01008128 CDW25489.1 GDIQ01258872 JAJ92852.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000007266 UP000235965 UP000053825 UP000053105 UP000005203 UP000242457 UP000053097 UP000279307 UP000008237 UP000005205 UP000078542 UP000075809 UP000007755 UP000078492 UP000078541 UP000009046 UP000078540 UP000192223 UP000000311 UP000002358 UP000015103 UP000019118 UP000007819 UP000245037 UP000027135 UP000030742 UP000092462 UP000092461 UP000075901 UP000075902 UP000075840 UP000075903 UP000075882 UP000076408 UP000215335 UP000075886 UP000075884 UP000075885 UP000076407 UP000075881 UP000075880 UP000075920 UP000030765 UP000075900 UP000008820 UP000069940 UP000249989 UP000069272 UP000036403 UP000075883 UP000183832 UP000000673 UP000078200 UP000002320 UP000092460 UP000092444 UP000092443 UP000007062 UP000091820 UP000009192 UP000008792 UP000092445 UP000001070
UP000007266 UP000235965 UP000053825 UP000053105 UP000005203 UP000242457 UP000053097 UP000279307 UP000008237 UP000005205 UP000078542 UP000075809 UP000007755 UP000078492 UP000078541 UP000009046 UP000078540 UP000192223 UP000000311 UP000002358 UP000015103 UP000019118 UP000007819 UP000245037 UP000027135 UP000030742 UP000092462 UP000092461 UP000075901 UP000075902 UP000075840 UP000075903 UP000075882 UP000076408 UP000215335 UP000075886 UP000075884 UP000075885 UP000076407 UP000075881 UP000075880 UP000075920 UP000030765 UP000075900 UP000008820 UP000069940 UP000249989 UP000069272 UP000036403 UP000075883 UP000183832 UP000000673 UP000078200 UP000002320 UP000092460 UP000092444 UP000092443 UP000007062 UP000091820 UP000009192 UP000008792 UP000092445 UP000001070
PRIDE
ProteinModelPortal
H9JQ60
A0A2A4J357
A0A2H1VFX9
A0A2A4J4B7
A0A2W1BHX7
A0A194QNX7
+ More
A0A194Q1N8 A0A212F7Y6 A0A0L7L1A7 D6WG56 A0A310SBM7 A0A2J7QAT2 A0A0L7QR14 A0A0N0U4I9 A0A087ZTV2 A0A2A3EGC3 A0A026X2F1 A0A2S2Q4W8 A0A3L8DSN7 E2C9Q5 A0A158P1F5 A0A195D127 A0A151WZB9 F4WYK3 A0A195E2S8 A0A195F3C3 E0VAT5 A0A151I2G3 A0A1W4WYZ4 E2AQE0 K7J869 T1HGT8 N6TWJ7 J9JQF1 A0A2P8XK82 A0A067R3Y2 U4U9V9 A0A1B6C7F8 A0A1B0CYH6 A0A1B0CRG5 A0A182SIP2 A0A182UK06 A0A182I7M5 A0A1S4H0N1 A0A2R7VZB5 A0A182VNS6 A0A182LME4 A0A182YFS5 A0A232EIB4 A0A182QRG8 A0A336L1F0 A0A182NBF0 A0A182PUW5 A0A182WXR2 A0A182KDD8 A0A182JJX8 A0A182VU18 A0A336L5G1 A0A084VKI4 A0A182RLC1 A0A1S4FYD2 Q16K05 A0A182H6D4 A0A182FNC9 A0A0J7NGC7 A0A182LYJ8 A0A2M3ZAA6 A0A1J1IKX8 W5J4U7 A0A1A9UTA7 B0XKL0 B0WXH2 A0A1B0AV56 A0A1B0FPX0 A0A2M3ZAV4 A0A1A9Y5Q7 A7UV83 A0A1A9WJ38 B4KHT4 A0A0Q9WJE5 A0A1A9ZM09 B4JPY0 A0A0K2THS8 A0A0P5F8U7
A0A194Q1N8 A0A212F7Y6 A0A0L7L1A7 D6WG56 A0A310SBM7 A0A2J7QAT2 A0A0L7QR14 A0A0N0U4I9 A0A087ZTV2 A0A2A3EGC3 A0A026X2F1 A0A2S2Q4W8 A0A3L8DSN7 E2C9Q5 A0A158P1F5 A0A195D127 A0A151WZB9 F4WYK3 A0A195E2S8 A0A195F3C3 E0VAT5 A0A151I2G3 A0A1W4WYZ4 E2AQE0 K7J869 T1HGT8 N6TWJ7 J9JQF1 A0A2P8XK82 A0A067R3Y2 U4U9V9 A0A1B6C7F8 A0A1B0CYH6 A0A1B0CRG5 A0A182SIP2 A0A182UK06 A0A182I7M5 A0A1S4H0N1 A0A2R7VZB5 A0A182VNS6 A0A182LME4 A0A182YFS5 A0A232EIB4 A0A182QRG8 A0A336L1F0 A0A182NBF0 A0A182PUW5 A0A182WXR2 A0A182KDD8 A0A182JJX8 A0A182VU18 A0A336L5G1 A0A084VKI4 A0A182RLC1 A0A1S4FYD2 Q16K05 A0A182H6D4 A0A182FNC9 A0A0J7NGC7 A0A182LYJ8 A0A2M3ZAA6 A0A1J1IKX8 W5J4U7 A0A1A9UTA7 B0XKL0 B0WXH2 A0A1B0AV56 A0A1B0FPX0 A0A2M3ZAV4 A0A1A9Y5Q7 A7UV83 A0A1A9WJ38 B4KHT4 A0A0Q9WJE5 A0A1A9ZM09 B4JPY0 A0A0K2THS8 A0A0P5F8U7
Ontologies
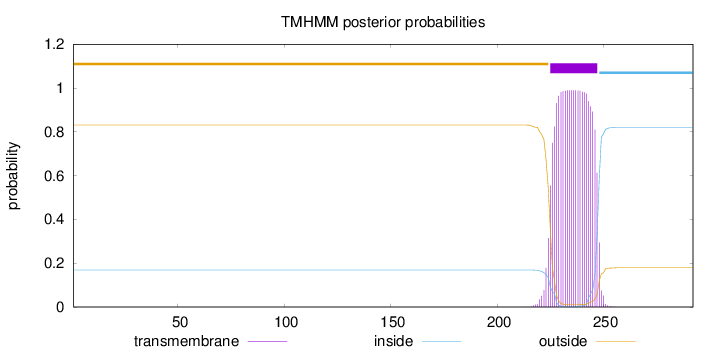
Topology
Length:
292
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.15176
Exp number, first 60 AAs:
0
Total prob of N-in:
0.16922
outside
1 - 224
TMhelix
225 - 247
inside
248 - 292
Population Genetic Test Statistics
Pi
192.599669
Theta
201.804529
Tajima's D
-0.808528
CLR
178.78948
CSRT
0.174041297935103
Interpretation
Uncertain
