Pre Gene Modal
BGIBMGA011659
Annotation
PREDICTED:_SRSF_protein_kinase_2_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 1.309 PlasmaMembrane Reliability : 1.67
Sequence
CDS
ATGCCTGAAAAGCAAAATTTTACAAAGAAAAGTATTTTAGTCACACCTAGCAGAGTTGTGAATAACCCAGATGAGATTGAAGGTGGAGAAGAAACTGAGATTGCTGCACAAATATCGGTTCCTCAAGATGGAGGCTGGGGATGGGTGGTCGTTATTGGAGCTTTTTTCAGCATTCTAATAATGGATGGAGTAGTATTTACATTTGGAAGCTTATTAAACGACATGGCGAACGATTTGAAATTGCAGGAATCCTTAGTGAATCTAATAAATTCAATCGCCGTAGCGTTATATTTTGTAATGGGTCCATTGGCTTCAGCATTGATCAATCGATTCGGTTTCCGGGCGTGTATGATGACTGGGTCTGTGATCTGTACGTTTTGTCTTTTTTGTTCATATTTTATTACGAGTTATTTAGGAATGTGTATATTTTATGGTGCAATGACTGGTTTTGGGTGTTGCTTAATTAATATGTCTTCAAGTTTGGTGGTTGGTTTTTATTTTGAGAAACTTCGATCAATTGCTTTGTCAGTGACTACAACAGGATCAAGCATTGGAATAATGATCATGTTTCCAATAAACTCCTATTTAGTCAAGTTAGGAGGATGGAGATTACCTACCCTAATACACTCCGGTTTATTCGGCTTAATATTTTACTTCGGTATGACATTTAGACCATTACTATCGCTGACTGTTGTTAATACTTCTAAATCTGATGTAAATAGAAGAGTACTCGCCATACAGGCGAAGAAAAAAAGACACAAAGTTGGTAAAAAGAAAAATCGCGAGGAAGAACACGGGGCTATTGGCCAAGGAAACGGTCTGCGTAACCAGAACCAGAATCGTCTAGAACCAACGCATAGTTCTTCCAACGAAACAATTGAGGAAGACGAAGATGAGCAATACACCAGCGACGACGAGGAGCAGGAGGATAGTGCGGATTACTGCAAGGGTGGATATCATCCTGTCAAGATTGGAGATTTATTCCTGAATCGTTACCACGTCACTCGGAAGCTAGGCTGGGGTCATTTTTCTACCGTGTGGTTATGCTGGGACCTCGTCGACAAACGATTCGTTGCGTTAAAAGTAGTGAAATCTGCACCTCATTTTACGGAGACGGCTTTGGACGAAATTAAAATATTGAGAGCAGTGAGAGACAGCGATCCCACCGATCCTAAGAGAAACAAAACCGTACAACTACTTAATGACTTTAAAATAACTGGTGTAAATGGCACTCATGTGTGCATGGTGTTTGAGGTTCTAGGACACCACCTTTTGAAATTGATACTCAGATCTAATTACCGAGGGATTCCTCGAGAGAATGTTAAGACGATAATTCGTCAGGTTTTAGAAGGCTTGGATTACCTTCACACTAAATGTAAAATTATTCATACTGACATTAAACCTGAAAATGTCCTTATCTGTGTAGATGAGACATATATACGTAAACTTGCCGCCGAAGCGACAGAATTACATTCCTTGGGGCTAAGGTTGCCTCATTCCCTCATTAGTACTGCTCCAAAAGAATTTCAAGAACAAGTGGTCACAGGTAAAATGAGCAGAAACAAGAAGAAAAAGTTGAAAAAGAAAGCTAAGCGTCAATCAATGTTGTTAAAGAAACAAATGGAACAAATAGAAGAAATGGAAGAACAAAAAAAGGATCCACACAATAGTGAATCTGCTCCTATGTCTCAAGATAATGATGATTCACCTCAGACACCAGAGGAAGCAAGTGTCATTGATATAACACAGTCTGAAAATGATACAGCAGTAAATGGGTGTAATGATGTCCAAGCTAATGAAGAAGAAACATATAACATTGATGGTATACAATTGCGAAACAAACAGTATGGGTGTGGTGATGATGCTAGCACTCCAATGTCCGAAGGAGAGATGACTGATACACCTCCTTCATTTAATTCACTTTCTACAAATCGTAAATCTTTAGACAAAAAGCCTGATCCTACTTTTGAAGTCTGTGACATTGAAGTAAAGATTGCGGATCTAGGTAATGCATGTTGGGTACATCGTCACTTTACTGAAGACATACAGACTAGGCAGTATCGTTCCTTAGAAGTGCTCCTAAGTGCTGGATATGGAACGTCAGCAGATATATGGAGCACAGCCTGTATGGCATTTGAATTAGCCACTGGAGATTATTTGTTTGAGCCTCACTCAGGTGATGGCTACAGTAGAGATGAGGATCACCTTGCTCATATTATTGAACTACTTGGAGATATTCCCAAAAGAATTGCAGCTTCAGGAAAATATTCCAAAGTATTTTTCAATAAAAAAGGTGAATTGCGAAATATAACAGGTTTAAAACCTTGGGGCTTAGTCTCTGTTCTGACAGAAAAATATGAATGGAGTGAAAATGATGCAGAAGAGTTTGCCGATTTTTTAAAACCTATGTTGGATTTTGATCCAAATCGGCGTGCCACTGCTTATGAGTGCCTTCAGCACCCATGGTTAAAATCGTCCAAACCAGAAAAGGAGTCTGAGGACTAA
Protein
MPEKQNFTKKSILVTPSRVVNNPDEIEGGEETEIAAQISVPQDGGWGWVVVIGAFFSILIMDGVVFTFGSLLNDMANDLKLQESLVNLINSIAVALYFVMGPLASALINRFGFRACMMTGSVICTFCLFCSYFITSYLGMCIFYGAMTGFGCCLINMSSSLVVGFYFEKLRSIALSVTTTGSSIGIMIMFPINSYLVKLGGWRLPTLIHSGLFGLIFYFGMTFRPLLSLTVVNTSKSDVNRRVLAIQAKKKRHKVGKKKNREEEHGAIGQGNGLRNQNQNRLEPTHSSSNETIEEDEDEQYTSDDEEQEDSADYCKGGYHPVKIGDLFLNRYHVTRKLGWGHFSTVWLCWDLVDKRFVALKVVKSAPHFTETALDEIKILRAVRDSDPTDPKRNKTVQLLNDFKITGVNGTHVCMVFEVLGHHLLKLILRSNYRGIPRENVKTIIRQVLEGLDYLHTKCKIIHTDIKPENVLICVDETYIRKLAAEATELHSLGLRLPHSLISTAPKEFQEQVVTGKMSRNKKKKLKKKAKRQSMLLKKQMEQIEEMEEQKKDPHNSESAPMSQDNDDSPQTPEEASVIDITQSENDTAVNGCNDVQANEEETYNIDGIQLRNKQYGCGDDASTPMSEGEMTDTPPSFNSLSTNRKSLDKKPDPTFEVCDIEVKIADLGNACWVHRHFTEDIQTRQYRSLEVLLSAGYGTSADIWSTACMAFELATGDYLFEPHSGDGYSRDEDHLAHIIELLGDIPKRIAASGKYSKVFFNKKGELRNITGLKPWGLVSVLTEKYEWSENDAEEFADFLKPMLDFDPNRRATAYECLQHPWLKSSKPEKESED
Summary
Uniprot
H9JQ53
A0A2A4K249
A0A2W1BPV1
A0A2H1WXW3
A0A194QPG5
A0A194PV75
+ More
A0A212ENU3 A0A0L7LBJ9 A0A2P8Y9P7 D6WGF8 R4WDC5 E2A1X4 A0A0J7KYX8 A0A026W7G3 A0A3L8DRJ5 A0A195BCJ8 A0A0M9ADM3 A0A087ZZL2 K7J6L4 E2B5T6 A0A1W4WLD0 A0A0L7R9H0 A0A146LMA3 A0A0A9ZEY4 A0A0T6B928 A0A1B6G6E1 A0A1B6LWQ2 A0A023FA63 A0A1J1IPP5 A0A1B6K5A1 N6TTC6 A0A1B6CLY6 A0A0P5CPA3 A0A1B6E3S5 A0A0P5BYD2 A0A0P5KDZ1 A0A0P5BDI3 A0A1Y1N9H8 A0A1L8DM64 A0A0P5KZX7 A0A1L8DMI5 A0A0N8E410 A0A0P5KWU5 A0A0P5HZF4 A0A0N8B5L6 A0A0N8B0M7 A0A0P5Z5V8 A0A0P4Y401 A0A2A3EK72 A0A0P4Y9S3 A0A0P4Y9B0 A0A0P5XNY2 A0A310S6W6 A0A0P4YTU3 A0A1B0CJI0 A0A0P4Z6Y4 A0A0P4ZER0
A0A212ENU3 A0A0L7LBJ9 A0A2P8Y9P7 D6WGF8 R4WDC5 E2A1X4 A0A0J7KYX8 A0A026W7G3 A0A3L8DRJ5 A0A195BCJ8 A0A0M9ADM3 A0A087ZZL2 K7J6L4 E2B5T6 A0A1W4WLD0 A0A0L7R9H0 A0A146LMA3 A0A0A9ZEY4 A0A0T6B928 A0A1B6G6E1 A0A1B6LWQ2 A0A023FA63 A0A1J1IPP5 A0A1B6K5A1 N6TTC6 A0A1B6CLY6 A0A0P5CPA3 A0A1B6E3S5 A0A0P5BYD2 A0A0P5KDZ1 A0A0P5BDI3 A0A1Y1N9H8 A0A1L8DM64 A0A0P5KZX7 A0A1L8DMI5 A0A0N8E410 A0A0P5KWU5 A0A0P5HZF4 A0A0N8B5L6 A0A0N8B0M7 A0A0P5Z5V8 A0A0P4Y401 A0A2A3EK72 A0A0P4Y9S3 A0A0P4Y9B0 A0A0P5XNY2 A0A310S6W6 A0A0P4YTU3 A0A1B0CJI0 A0A0P4Z6Y4 A0A0P4ZER0
Pubmed
EMBL
BABH01012118
NWSH01000215
PCG78317.1
KZ150089
PZC73713.1
ODYU01011901
+ More
SOQ57910.1 KQ461190 KPJ07244.1 KQ459590 KPI97301.1 AGBW02013617 OWR43146.1 JTDY01001830 KOB72770.1 PYGN01000773 PSN40987.1 KQ971321 EFA00182.1 AK417553 BAN20768.1 GL435845 EFN72568.1 LBMM01001942 KMQ95503.1 KK107361 EZA51995.1 QOIP01000005 RLU23065.1 KQ976514 KYM82291.1 KQ435691 KOX81153.1 AAZX01003985 AAZX01013517 GL445887 EFN88905.1 KQ414627 KOC67401.1 GDHC01010314 JAQ08315.1 GBHO01000590 GBRD01009908 JAG43014.1 JAG55916.1 LJIG01009069 KRT83817.1 GECZ01011775 JAS57994.1 GEBQ01011854 JAT28123.1 GBBI01000818 JAC17894.1 CVRI01000054 CRL00473.1 GECU01001088 JAT06619.1 APGK01055188 KB741261 KB632254 ENN71596.1 ERL90624.1 GEDC01030842 GEDC01029826 GEDC01024689 GEDC01023080 GEDC01022997 GEDC01004363 GEDC01001203 JAS06456.1 JAS07472.1 JAS12609.1 JAS14218.1 JAS14301.1 JAS32935.1 JAS36095.1 GDIP01183411 JAJ39991.1 GEDC01004705 JAS32593.1 GDIP01182166 JAJ41236.1 GDIQ01185545 JAK66180.1 GDIP01186060 JAJ37342.1 GEZM01013963 JAV92247.1 GFDF01006535 JAV07549.1 GDIQ01185544 JAK66181.1 GFDF01006534 JAV07550.1 GDIQ01063988 JAN30749.1 GDIQ01178948 JAK72777.1 GDIQ01243536 JAK08189.1 GDIQ01210531 JAK41194.1 GDIQ01224443 JAK27282.1 GDIP01048580 JAM55135.1 GDIP01233016 JAI90385.1 KZ288230 PBC31579.1 GDIP01230722 JAI92679.1 GDIP01233015 JAI90386.1 GDIP01071736 JAM31979.1 KQ767065 OAD53514.1 GDIP01222517 JAJ00885.1 AJWK01014662 GDIP01221371 JAJ02031.1 GDIP01214567 JAJ08835.1
SOQ57910.1 KQ461190 KPJ07244.1 KQ459590 KPI97301.1 AGBW02013617 OWR43146.1 JTDY01001830 KOB72770.1 PYGN01000773 PSN40987.1 KQ971321 EFA00182.1 AK417553 BAN20768.1 GL435845 EFN72568.1 LBMM01001942 KMQ95503.1 KK107361 EZA51995.1 QOIP01000005 RLU23065.1 KQ976514 KYM82291.1 KQ435691 KOX81153.1 AAZX01003985 AAZX01013517 GL445887 EFN88905.1 KQ414627 KOC67401.1 GDHC01010314 JAQ08315.1 GBHO01000590 GBRD01009908 JAG43014.1 JAG55916.1 LJIG01009069 KRT83817.1 GECZ01011775 JAS57994.1 GEBQ01011854 JAT28123.1 GBBI01000818 JAC17894.1 CVRI01000054 CRL00473.1 GECU01001088 JAT06619.1 APGK01055188 KB741261 KB632254 ENN71596.1 ERL90624.1 GEDC01030842 GEDC01029826 GEDC01024689 GEDC01023080 GEDC01022997 GEDC01004363 GEDC01001203 JAS06456.1 JAS07472.1 JAS12609.1 JAS14218.1 JAS14301.1 JAS32935.1 JAS36095.1 GDIP01183411 JAJ39991.1 GEDC01004705 JAS32593.1 GDIP01182166 JAJ41236.1 GDIQ01185545 JAK66180.1 GDIP01186060 JAJ37342.1 GEZM01013963 JAV92247.1 GFDF01006535 JAV07549.1 GDIQ01185544 JAK66181.1 GFDF01006534 JAV07550.1 GDIQ01063988 JAN30749.1 GDIQ01178948 JAK72777.1 GDIQ01243536 JAK08189.1 GDIQ01210531 JAK41194.1 GDIQ01224443 JAK27282.1 GDIP01048580 JAM55135.1 GDIP01233016 JAI90385.1 KZ288230 PBC31579.1 GDIP01230722 JAI92679.1 GDIP01233015 JAI90386.1 GDIP01071736 JAM31979.1 KQ767065 OAD53514.1 GDIP01222517 JAJ00885.1 AJWK01014662 GDIP01221371 JAJ02031.1 GDIP01214567 JAJ08835.1
Proteomes
PRIDE
Interpro
CDD
ProteinModelPortal
H9JQ53
A0A2A4K249
A0A2W1BPV1
A0A2H1WXW3
A0A194QPG5
A0A194PV75
+ More
A0A212ENU3 A0A0L7LBJ9 A0A2P8Y9P7 D6WGF8 R4WDC5 E2A1X4 A0A0J7KYX8 A0A026W7G3 A0A3L8DRJ5 A0A195BCJ8 A0A0M9ADM3 A0A087ZZL2 K7J6L4 E2B5T6 A0A1W4WLD0 A0A0L7R9H0 A0A146LMA3 A0A0A9ZEY4 A0A0T6B928 A0A1B6G6E1 A0A1B6LWQ2 A0A023FA63 A0A1J1IPP5 A0A1B6K5A1 N6TTC6 A0A1B6CLY6 A0A0P5CPA3 A0A1B6E3S5 A0A0P5BYD2 A0A0P5KDZ1 A0A0P5BDI3 A0A1Y1N9H8 A0A1L8DM64 A0A0P5KZX7 A0A1L8DMI5 A0A0N8E410 A0A0P5KWU5 A0A0P5HZF4 A0A0N8B5L6 A0A0N8B0M7 A0A0P5Z5V8 A0A0P4Y401 A0A2A3EK72 A0A0P4Y9S3 A0A0P4Y9B0 A0A0P5XNY2 A0A310S6W6 A0A0P4YTU3 A0A1B0CJI0 A0A0P4Z6Y4 A0A0P4ZER0
A0A212ENU3 A0A0L7LBJ9 A0A2P8Y9P7 D6WGF8 R4WDC5 E2A1X4 A0A0J7KYX8 A0A026W7G3 A0A3L8DRJ5 A0A195BCJ8 A0A0M9ADM3 A0A087ZZL2 K7J6L4 E2B5T6 A0A1W4WLD0 A0A0L7R9H0 A0A146LMA3 A0A0A9ZEY4 A0A0T6B928 A0A1B6G6E1 A0A1B6LWQ2 A0A023FA63 A0A1J1IPP5 A0A1B6K5A1 N6TTC6 A0A1B6CLY6 A0A0P5CPA3 A0A1B6E3S5 A0A0P5BYD2 A0A0P5KDZ1 A0A0P5BDI3 A0A1Y1N9H8 A0A1L8DM64 A0A0P5KZX7 A0A1L8DMI5 A0A0N8E410 A0A0P5KWU5 A0A0P5HZF4 A0A0N8B5L6 A0A0N8B0M7 A0A0P5Z5V8 A0A0P4Y401 A0A2A3EK72 A0A0P4Y9S3 A0A0P4Y9B0 A0A0P5XNY2 A0A310S6W6 A0A0P4YTU3 A0A1B0CJI0 A0A0P4Z6Y4 A0A0P4ZER0
PDB
5MYV
E-value=1.80074e-92,
Score=869
Ontologies
GO
Topology
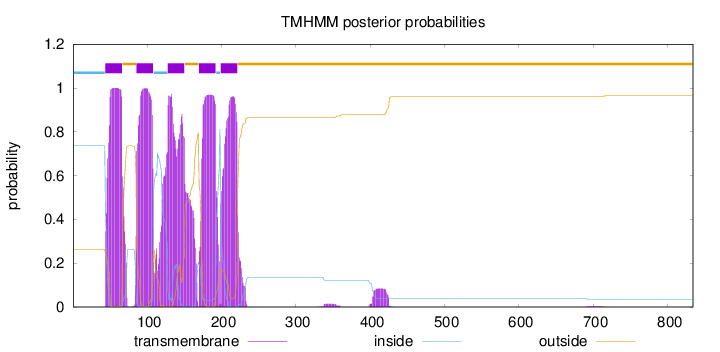
Length:
834
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
123.59856
Exp number, first 60 AAs:
14.5679
Total prob of N-in:
0.73684
POSSIBLE N-term signal
sequence
inside
1 - 43
TMhelix
44 - 66
outside
67 - 85
TMhelix
86 - 108
inside
109 - 127
TMhelix
128 - 150
outside
151 - 169
TMhelix
170 - 192
inside
193 - 198
TMhelix
199 - 221
outside
222 - 834
Population Genetic Test Statistics
Pi
303.578593
Theta
232.305089
Tajima's D
1.003873
CLR
0
CSRT
0.661166941652917
Interpretation
Uncertain
