Pre Gene Modal
BGIBMGA011818
Annotation
PREDICTED:_DNA_replication_factor_Cdt1_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.803
Sequence
CDS
ATGTCCCAAACAACGATTACATCATTTTATAACAGCAGAAAAAGACCAGCAGCGGATGATATAGCCACAACCAAGAGTAAAATTGCACATTTAGACCGTGCGTTGGAATCTAAAGCACTTAATAAAGCTACGGCTGCTAAGAAAACAGAAACGGTAATTGATATTAAGTTAACAAATGATGATATCAAGACTGATGTAAATACTGATATAACATCACCAGCTCCAAAAGTGCCCGAAAAATGTAAAGAAAAAGTAGCTTTTACGAGGAAGGGAAATGTGCCCAAGACAACTCAATTGTCACAACGTGAAACTATAAAATCAGCTCGACAGGAACTAAGCCTAGGAGACATTAGGAAAAAATTAGCTAATAGTTCGAAGCTTGCAGAACTGAAAGCATCTGCTGAACGTATTAGCAGAGGTATCAAGCTATTAAAAGAATCCAATGAGACAAAGAATCTAAAAGAATTCAAGTCTATCAATGTGGAAGTACCCTCGAGTCCAAGTAAGAGAGTTCCAGTAAGAAATGAATTGCTTCCTCAAAAAGCTGATGTTCCTATAAGTCCTCAAAACTCTCTCATTTCTCCAAGAAAAGTTTTTGTGAGTCCAGTGAAGAGCCCTAGCAAGGTACCAGCGTATATCAGACATGCATCATTGGCAACAACTTCTACTAGCCTGCCATTACCGCATAAATATAGGTTCCTGGCTGAGCTATTCAGAGGTATGGAGACAGTTGTGGCATTGCTTTATAACAGAAATGAGAAGATAACATTCACAAAACTCAAACCATCAGTCCAGGAAATGCTCAAAAGAAACTTTGGAGAGAAACATTTGGCACAAATAAAGCACTTAGTGCCAGATTTTTACAATTTTGAAGTACAAAAGATTAAAAACTTCAGTACATCTTCTCATAAAGATGCCTATGAACTTGTTGTAACTCCAAATTTTCCCAATGATATAAAAAATATGAATCCAAGTGTGCTTCTGGAGAGGAGAAGGTTTTTCTTTAATACCCTTTTGCAGGTAGTGATGAAACATCATTCTCAATATCTCCTTACTCTTGACCCTCCAATGGTCATACCAGATAATAAGTTAACACGTTGGCATCCAGAATTTGAGTTGGAGAAATTGCCTGAAATTGAAAGTGCTAAATTGCCTGAGCTGCCAAATGTAGAAAAAATGTCAACAGCTCAAGATGTACTAGCTAAAGCACGTGAACTATTCAAATGTAATACTAAAATGGAGAGAGCTATGGAGAAATTAGCACACGCTAAAGCACGGGGCCTAACTGAGCAGGAGAAATTAGTTACTGGCATTACAGATGATAACAAAAAAAACACTGTTACTAATGGAACTCAAACTCCTACTACAAGCACAATGACACTTCTCAATCCTGCCTTGCGTAATTTACCAACAGCTCTCTTGGAGAAAGTTAAAGCTAAGCAGGCAGCCAAAGCACTCGAGGCCATGACAAGGTCTTCAGAAACTGAACAAAAATATCACATTTACAGTAGACTTCCAGATCTCGCCAGAACACTAAGAAACATTTTTGTCACTGAAAGAAAGAATGTCCTTGCATTGAACATTATTCTATCTAAATTAGACAGCAGTTTTAAATCTAAGGTATCAGCCAATGATTTACATAGAGACATAAAAATTCTGACAGACGAAGTTCCTGAATGGTGTAAATTGCATGAAATAAGAAATACTTCATATTTGAAATTGGATCGAAATTCAGACTTAAACACAATTATCTGTAAATTAGAAGCTTTGGCTGATAAATACAAAGTAGACTAA
Protein
MSQTTITSFYNSRKRPAADDIATTKSKIAHLDRALESKALNKATAAKKTETVIDIKLTNDDIKTDVNTDITSPAPKVPEKCKEKVAFTRKGNVPKTTQLSQRETIKSARQELSLGDIRKKLANSSKLAELKASAERISRGIKLLKESNETKNLKEFKSINVEVPSSPSKRVPVRNELLPQKADVPISPQNSLISPRKVFVSPVKSPSKVPAYIRHASLATTSTSLPLPHKYRFLAELFRGMETVVALLYNRNEKITFTKLKPSVQEMLKRNFGEKHLAQIKHLVPDFYNFEVQKIKNFSTSSHKDAYELVVTPNFPNDIKNMNPSVLLERRRFFFNTLLQVVMKHHSQYLLTLDPPMVIPDNKLTRWHPEFELEKLPEIESAKLPELPNVEKMSTAQDVLAKARELFKCNTKMERAMEKLAHAKARGLTEQEKLVTGITDDNKKNTVTNGTQTPTTSTMTLLNPALRNLPTALLEKVKAKQAAKALEAMTRSSETEQKYHIYSRLPDLARTLRNIFVTERKNVLALNIILSKLDSSFKSKVSANDLHRDIKILTDEVPEWCKLHEIRNTSYLKLDRNSDLNTIICKLEALADKYKVD
Summary
Uniprot
L0L004
H7CHT8
A0A2W1BHG6
A0A2A4K2P4
A0A2H1WXY0
A0A0L7LIF2
+ More
A0A212ENT5 S4NSY3 A0A194PX09 A0A194QUL4 A0A0C9R7R8 A0A154P0Q6 A0A2A3ES10 A0A0J7KS21 A0A088A579 A0A034WI11 A0A0K8WCJ3 A0A0L7QXA1 T1PJP0 A0A026WAM5 A0A151I190 A0A1I8MFM8 E9IVA5 A0A195DJ21 A0A195FJ23 A0A1I8NSJ8 A0A151XHY7 F4WSP4 D6WGF9 A0A0L0CEX0 A0A2J7Q8P3 A0A195C0G5 B4KM56 A0A1Q3EVB5 A0A1W4WZ13 K7ILZ6 A0A0A1X950 A0A232F8G1 A0A310SLQ5 A0A0N0U821 A0A1S4FJM9 A0A182GCH9 A0A182GF11 A0A0K8TND6 A0A1Y1L6K2 B4J5Q3 B0WRV5 Q16YK2 A0A0P6JSN4 E2C608 B3MEW7 A0A0M4EV13 A0A1B0G327 A0A1A9V8Y6 U4UKU4 A0A1B0BRB7 A0A1A9ZF06 A0A1A9Y5I0 N6TQW3 A0A0M5J9S2 E2BND6 E2AFD3 A0A1A9W7N2 A0A336MXV3 B4LP18 A0A182RA20 B4NMV2 A0A336MBD5 A0A067QKA3 U5EZH4 A0A084W2H5 A0A1B6CU63 A0A2P8YA04 A0A1A9XP84 A0A182JQA0 W5JT66 A0A182FCD6 A0A182V5W6 A0A1B0CJI1 A0A1A9Z596 A0A1B0GBN5 A0A182YFP1 A0A2M4CZ67 A0A182WB57 A0A182N876 A0A182XD49 A0A182KVY7 A0A182TIA0 A0A182QKE6 Q7PNP7 A0A182HMI4 A0A182PGG4 A0A1A9WV46 A0A0P4VMN2 A0A1B0CHV7 A0A1B0FFD9
A0A212ENT5 S4NSY3 A0A194PX09 A0A194QUL4 A0A0C9R7R8 A0A154P0Q6 A0A2A3ES10 A0A0J7KS21 A0A088A579 A0A034WI11 A0A0K8WCJ3 A0A0L7QXA1 T1PJP0 A0A026WAM5 A0A151I190 A0A1I8MFM8 E9IVA5 A0A195DJ21 A0A195FJ23 A0A1I8NSJ8 A0A151XHY7 F4WSP4 D6WGF9 A0A0L0CEX0 A0A2J7Q8P3 A0A195C0G5 B4KM56 A0A1Q3EVB5 A0A1W4WZ13 K7ILZ6 A0A0A1X950 A0A232F8G1 A0A310SLQ5 A0A0N0U821 A0A1S4FJM9 A0A182GCH9 A0A182GF11 A0A0K8TND6 A0A1Y1L6K2 B4J5Q3 B0WRV5 Q16YK2 A0A0P6JSN4 E2C608 B3MEW7 A0A0M4EV13 A0A1B0G327 A0A1A9V8Y6 U4UKU4 A0A1B0BRB7 A0A1A9ZF06 A0A1A9Y5I0 N6TQW3 A0A0M5J9S2 E2BND6 E2AFD3 A0A1A9W7N2 A0A336MXV3 B4LP18 A0A182RA20 B4NMV2 A0A336MBD5 A0A067QKA3 U5EZH4 A0A084W2H5 A0A1B6CU63 A0A2P8YA04 A0A1A9XP84 A0A182JQA0 W5JT66 A0A182FCD6 A0A182V5W6 A0A1B0CJI1 A0A1A9Z596 A0A1B0GBN5 A0A182YFP1 A0A2M4CZ67 A0A182WB57 A0A182N876 A0A182XD49 A0A182KVY7 A0A182TIA0 A0A182QKE6 Q7PNP7 A0A182HMI4 A0A182PGG4 A0A1A9WV46 A0A0P4VMN2 A0A1B0CHV7 A0A1B0FFD9
Pubmed
19121390
24773378
28756777
26227816
22118469
23622113
+ More
26354079 25348373 24508170 30249741 25315136 21282665 21719571 18362917 19820115 26108605 17994087 20075255 25830018 28648823 17510324 26483478 26369729 28004739 26999592 20798317 23537049 24845553 24438588 29403074 20920257 23761445 25244985 20966253 12364791 14747013 17210077 27129103
26354079 25348373 24508170 30249741 25315136 21282665 21719571 18362917 19820115 26108605 17994087 20075255 25830018 28648823 17510324 26483478 26369729 28004739 26999592 20798317 23537049 24845553 24438588 29403074 20920257 23761445 25244985 20966253 12364791 14747013 17210077 27129103
EMBL
JX069947
AGB51151.1
BABH01012118
BABH01012119
AB703260
BAL72669.1
+ More
KZ150089 PZC73711.1 NWSH01000215 PCG78319.1 ODYU01011901 SOQ57908.1 JTDY01000943 KOB75338.1 AGBW02013617 OWR43144.1 GAIX01013857 JAA78703.1 KQ459590 KPI97299.1 KQ461190 KPJ07246.1 GBYB01002789 GBYB01002791 JAG72556.1 JAG72558.1 KQ434783 KZC04828.1 KZ288191 PBC34480.1 LBMM01003750 KMQ93182.1 GAKP01005534 JAC53418.1 GDHF01003565 JAI48749.1 KQ414705 KOC63242.1 KA648909 AFP63538.1 KK107293 QOIP01000005 EZA53127.1 RLU22857.1 KQ976599 KYM79507.1 GL766194 EFZ15475.1 KQ980800 KYN12817.1 KQ981560 KYN39984.1 KQ982130 KYQ59830.1 GL888326 EGI62787.1 KQ971321 EFA00549.1 JRES01000480 KNC30938.1 NEVH01016949 PNF24958.1 KQ978379 KYM94394.1 CH933808 EDW08720.1 GFDL01015798 JAV19247.1 GBXI01006856 GBXI01006637 JAD07436.1 JAD07655.1 NNAY01000772 OXU26597.1 KQ759893 OAD62074.1 KQ435687 KOX81352.1 JXUM01054280 KQ561814 KXJ77444.1 JXUM01059063 KQ562033 KXJ76852.1 GDAI01001960 JAI15643.1 GEZM01063212 JAV69194.1 CH916367 EDW01829.1 DS232061 EDS33520.1 CH477515 EAT39705.1 GDUN01000248 JAN95671.1 GL452895 EFN76613.1 CH902619 EDV36588.1 CP012524 ALC41382.1 CCAG010009842 KB632254 ERL90625.1 JXJN01019030 APGK01055188 KB741261 ENN71595.1 ALC40431.1 GL449414 EFN82715.1 GL439108 EFN67826.1 UFQT01002460 SSX33513.1 CH940648 EDW62212.1 CH964282 EDW85691.1 UFQT01000700 SSX26641.1 KK853239 KDR09483.1 GANO01000791 JAB59080.1 ATLV01019619 KE525275 KFB44419.1 GEDC01020262 JAS17036.1 PYGN01000773 PSN40984.1 ADMH02000550 ETN65919.1 AJWK01014662 CCAG010005856 GGFL01006387 MBW70565.1 AXCN02001004 AAAB01008960 EAA11814.5 APCN01004443 GDKW01000173 JAI56422.1 AJWK01012745 CCAG010009702
KZ150089 PZC73711.1 NWSH01000215 PCG78319.1 ODYU01011901 SOQ57908.1 JTDY01000943 KOB75338.1 AGBW02013617 OWR43144.1 GAIX01013857 JAA78703.1 KQ459590 KPI97299.1 KQ461190 KPJ07246.1 GBYB01002789 GBYB01002791 JAG72556.1 JAG72558.1 KQ434783 KZC04828.1 KZ288191 PBC34480.1 LBMM01003750 KMQ93182.1 GAKP01005534 JAC53418.1 GDHF01003565 JAI48749.1 KQ414705 KOC63242.1 KA648909 AFP63538.1 KK107293 QOIP01000005 EZA53127.1 RLU22857.1 KQ976599 KYM79507.1 GL766194 EFZ15475.1 KQ980800 KYN12817.1 KQ981560 KYN39984.1 KQ982130 KYQ59830.1 GL888326 EGI62787.1 KQ971321 EFA00549.1 JRES01000480 KNC30938.1 NEVH01016949 PNF24958.1 KQ978379 KYM94394.1 CH933808 EDW08720.1 GFDL01015798 JAV19247.1 GBXI01006856 GBXI01006637 JAD07436.1 JAD07655.1 NNAY01000772 OXU26597.1 KQ759893 OAD62074.1 KQ435687 KOX81352.1 JXUM01054280 KQ561814 KXJ77444.1 JXUM01059063 KQ562033 KXJ76852.1 GDAI01001960 JAI15643.1 GEZM01063212 JAV69194.1 CH916367 EDW01829.1 DS232061 EDS33520.1 CH477515 EAT39705.1 GDUN01000248 JAN95671.1 GL452895 EFN76613.1 CH902619 EDV36588.1 CP012524 ALC41382.1 CCAG010009842 KB632254 ERL90625.1 JXJN01019030 APGK01055188 KB741261 ENN71595.1 ALC40431.1 GL449414 EFN82715.1 GL439108 EFN67826.1 UFQT01002460 SSX33513.1 CH940648 EDW62212.1 CH964282 EDW85691.1 UFQT01000700 SSX26641.1 KK853239 KDR09483.1 GANO01000791 JAB59080.1 ATLV01019619 KE525275 KFB44419.1 GEDC01020262 JAS17036.1 PYGN01000773 PSN40984.1 ADMH02000550 ETN65919.1 AJWK01014662 CCAG010005856 GGFL01006387 MBW70565.1 AXCN02001004 AAAB01008960 EAA11814.5 APCN01004443 GDKW01000173 JAI56422.1 AJWK01012745 CCAG010009702
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000053240
+ More
UP000076502 UP000242457 UP000036403 UP000005203 UP000053825 UP000053097 UP000279307 UP000078540 UP000095301 UP000078492 UP000078541 UP000095300 UP000075809 UP000007755 UP000007266 UP000037069 UP000235965 UP000078542 UP000009192 UP000192223 UP000002358 UP000215335 UP000053105 UP000069940 UP000249989 UP000001070 UP000002320 UP000008820 UP000008237 UP000007801 UP000092553 UP000092444 UP000078200 UP000030742 UP000092460 UP000092445 UP000092443 UP000019118 UP000000311 UP000091820 UP000008792 UP000075900 UP000007798 UP000027135 UP000030765 UP000245037 UP000075881 UP000000673 UP000069272 UP000075903 UP000092461 UP000076408 UP000075920 UP000075884 UP000076407 UP000075882 UP000075902 UP000075886 UP000007062 UP000075840 UP000075885
UP000076502 UP000242457 UP000036403 UP000005203 UP000053825 UP000053097 UP000279307 UP000078540 UP000095301 UP000078492 UP000078541 UP000095300 UP000075809 UP000007755 UP000007266 UP000037069 UP000235965 UP000078542 UP000009192 UP000192223 UP000002358 UP000215335 UP000053105 UP000069940 UP000249989 UP000001070 UP000002320 UP000008820 UP000008237 UP000007801 UP000092553 UP000092444 UP000078200 UP000030742 UP000092460 UP000092445 UP000092443 UP000019118 UP000000311 UP000091820 UP000008792 UP000075900 UP000007798 UP000027135 UP000030765 UP000245037 UP000075881 UP000000673 UP000069272 UP000075903 UP000092461 UP000076408 UP000075920 UP000075884 UP000076407 UP000075882 UP000075902 UP000075886 UP000007062 UP000075840 UP000075885
Interpro
SUPFAM
SSF46785
SSF46785
ProteinModelPortal
L0L004
H7CHT8
A0A2W1BHG6
A0A2A4K2P4
A0A2H1WXY0
A0A0L7LIF2
+ More
A0A212ENT5 S4NSY3 A0A194PX09 A0A194QUL4 A0A0C9R7R8 A0A154P0Q6 A0A2A3ES10 A0A0J7KS21 A0A088A579 A0A034WI11 A0A0K8WCJ3 A0A0L7QXA1 T1PJP0 A0A026WAM5 A0A151I190 A0A1I8MFM8 E9IVA5 A0A195DJ21 A0A195FJ23 A0A1I8NSJ8 A0A151XHY7 F4WSP4 D6WGF9 A0A0L0CEX0 A0A2J7Q8P3 A0A195C0G5 B4KM56 A0A1Q3EVB5 A0A1W4WZ13 K7ILZ6 A0A0A1X950 A0A232F8G1 A0A310SLQ5 A0A0N0U821 A0A1S4FJM9 A0A182GCH9 A0A182GF11 A0A0K8TND6 A0A1Y1L6K2 B4J5Q3 B0WRV5 Q16YK2 A0A0P6JSN4 E2C608 B3MEW7 A0A0M4EV13 A0A1B0G327 A0A1A9V8Y6 U4UKU4 A0A1B0BRB7 A0A1A9ZF06 A0A1A9Y5I0 N6TQW3 A0A0M5J9S2 E2BND6 E2AFD3 A0A1A9W7N2 A0A336MXV3 B4LP18 A0A182RA20 B4NMV2 A0A336MBD5 A0A067QKA3 U5EZH4 A0A084W2H5 A0A1B6CU63 A0A2P8YA04 A0A1A9XP84 A0A182JQA0 W5JT66 A0A182FCD6 A0A182V5W6 A0A1B0CJI1 A0A1A9Z596 A0A1B0GBN5 A0A182YFP1 A0A2M4CZ67 A0A182WB57 A0A182N876 A0A182XD49 A0A182KVY7 A0A182TIA0 A0A182QKE6 Q7PNP7 A0A182HMI4 A0A182PGG4 A0A1A9WV46 A0A0P4VMN2 A0A1B0CHV7 A0A1B0FFD9
A0A212ENT5 S4NSY3 A0A194PX09 A0A194QUL4 A0A0C9R7R8 A0A154P0Q6 A0A2A3ES10 A0A0J7KS21 A0A088A579 A0A034WI11 A0A0K8WCJ3 A0A0L7QXA1 T1PJP0 A0A026WAM5 A0A151I190 A0A1I8MFM8 E9IVA5 A0A195DJ21 A0A195FJ23 A0A1I8NSJ8 A0A151XHY7 F4WSP4 D6WGF9 A0A0L0CEX0 A0A2J7Q8P3 A0A195C0G5 B4KM56 A0A1Q3EVB5 A0A1W4WZ13 K7ILZ6 A0A0A1X950 A0A232F8G1 A0A310SLQ5 A0A0N0U821 A0A1S4FJM9 A0A182GCH9 A0A182GF11 A0A0K8TND6 A0A1Y1L6K2 B4J5Q3 B0WRV5 Q16YK2 A0A0P6JSN4 E2C608 B3MEW7 A0A0M4EV13 A0A1B0G327 A0A1A9V8Y6 U4UKU4 A0A1B0BRB7 A0A1A9ZF06 A0A1A9Y5I0 N6TQW3 A0A0M5J9S2 E2BND6 E2AFD3 A0A1A9W7N2 A0A336MXV3 B4LP18 A0A182RA20 B4NMV2 A0A336MBD5 A0A067QKA3 U5EZH4 A0A084W2H5 A0A1B6CU63 A0A2P8YA04 A0A1A9XP84 A0A182JQA0 W5JT66 A0A182FCD6 A0A182V5W6 A0A1B0CJI1 A0A1A9Z596 A0A1B0GBN5 A0A182YFP1 A0A2M4CZ67 A0A182WB57 A0A182N876 A0A182XD49 A0A182KVY7 A0A182TIA0 A0A182QKE6 Q7PNP7 A0A182HMI4 A0A182PGG4 A0A1A9WV46 A0A0P4VMN2 A0A1B0CHV7 A0A1B0FFD9
PDB
2WVR
E-value=8.04836e-59,
Score=577
Ontologies
GO
Topology
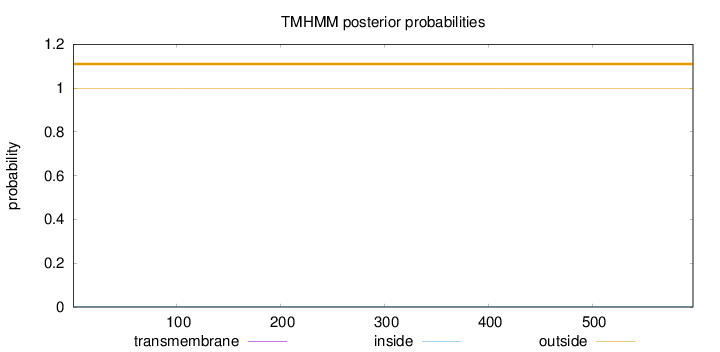
Length:
597
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000740000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00136
outside
1 - 597
Population Genetic Test Statistics
Pi
266.014181
Theta
152.546789
Tajima's D
-1.665997
CLR
103.356973
CSRT
0.0420478976051197
Interpretation
Uncertain
