Gene
KWMTBOMO06564
Pre Gene Modal
BGIBMGA011657
Annotation
PREDICTED:_lisH_domain_and_HEAT_repeat-containing_protein_KIAA1468-like_[Bombyx_mori]
Location in the cell
Golgi Reliability : 1.151 Nuclear Reliability : 1.003 PlasmaMembrane Reliability : 1.447
Sequence
CDS
ATGAATCCGTACAAGGAAACAGAGGAGTCATCTTTCGATACACCTCATTTGTCTTATGAGGATATAGCTACAAAACTGCTAAAAGACAACTTCTTCCTGACTGCCTTAGAACTCCACACTGAGCTTGTAGAGAGCGGCAAGGAACTTCCTCAACTTAGAGAATTCTTCTCGAATCCTGGAAATTTTGAACAGCATGTCTCTAGAGCATCTGAATTAAATTCTATACACCGCACTCCAAGTCAAGCAACTTTAGACTCTTTGGACACAGCAAGATATTCAGAAGATGGTGGAGGTGACCGTGGTGGTAGTGGGGGTGACGTGGCTGTTCTAGAATTTGAGCTGCGTAAGGCAAGGGAAACTATTAATTCCCTAAGGGCTAATCTTACACAGTTTGCAGATGGTTCATCAACATTAGATAAAAGTTCCCAAAAAATATGTGACCAGAGATCCTTAAAGCCACATGAGAAGAGAGCACTCAATTTCTTGATTAATGAATATCTACTTATGCAAGATTATAAATTAACTTCAATTACATTTTCTGATGAGAATCCTGATCAGGAATTCGAAGATTGGGATGATGTAGGTTTGAATATGCCTCGACCCGCAGGAATAGTCTCCCTATTCCGAGGAGAGTCAGTATTGAGTGTACCTAAAGTAGACTCATCAACTGAAACAAATTGGCAACAAATAGATGCTGGATGCCAGACTGAACTTGATGAAAGGAGTATTTGTGTCAGTTGCCAGGACTCGATTGATCATGATAATGATTGGGCAGCTGAGCTTCTCATTCAAGCTGAAGAGATAGAAATGCTGAAACAAAAGGCTATGGCCTTAGAGATGGAGAAGCAAAATTTTCAAAAATTGTATGATACTGCTATTGTCAGTTTGAATTCTCTTGCAAGTCCCGCCATCGAGAGCAAAGCCCTTGATTTAAACACAAAAGATGAAAAAATGATTGAAGCCGAACAGGCTATTGATACTACACCAATAACTTGTACGGCTTCAGAGAATCATTACGGGAGTGGAAATTCTACACATAGTGCAACACCAGAACAGTTCGAAATGATTGAGAGTGAAAAGAATAGTACTATCACCAGAAAAGAAGGCTCTACGCTAAGTTCCTTTGAACATACTGTGTTGGGTGAAAATTGTAACAGAGATTCACCGTTGCGGCGCGGTAGTGTCACCACTTTAGACGAAGCCCTTAGCATAAATGATGGAGGCGAGTGGACACGTGTTCAATATGAATACAACAGCATCGAAACTAGTAGTAAGGAAATGTGGATCGACAGTGGTCTACCAAATAGTCTAAAAGTATCAATTCTAAACTGGTGCTGTGAGACGCTGAACCTGAACGAATCATTCTCGAACGATCTTTTAGTAGAACTAATAAACAATGATAAACCAATAACACTGCCTGGGTTTCTAACGCTGGCTGCCGACACTATGCCTAGGATAATCCCTCATACTCTCGTGGCACGACGTTTCGAAGCTGCGGCATTACTAGCAGCGAGCGTGGCTCTGCTCCCGACCGGAGATCCAAGACGCGCTCGCACCTTACACTCATTACTTACACTCGTGAAGAAGCCGGAGCCGTCCGACACGAGATCCATATGTGAAGCGGCGTGTTACATAGTGAAATGGGGAGGCAGCGGGGAAGTGTTGTCCTCCGTAGCTGAATTACTGGCATCGAGATCGGCCGAGCGACGGATCCTCGCCAGCCAGATCTGCGTCGCCATAGCTCCGTACGTTCCGATCGAGCTGTGCACGTCCCTACTGCTCAGTCTAGTGGTGCTGATGTGCGAATCGAACGAGATCGAAATCAGGAGTTTGGCCCTAAAATCCGCGTGCCTAATATGTCCGGTTGCGGAACATAAGTACGCGCAGCTCGAGAATATAATGTTCGGTTTCCTGAAGGATCCGATCGAGAAGCACGTCAAAGATACGGTCAGCATTTTCGGGCCGATACTGGCGAGAAGCGCTCTGATGGCAGGAAAATTCTGTACCGATCTGTGTACACGTATTATGAGAAACGCTGCGAAAGAGAGCTCGGATAATGAATGGAAAAACGCTGCTCTGTATTTAGATCTTATGAAATCTTTAGTATTAGCGAAACTGGCATACGTTATAAACGTACAAATAATCAAAGATGCCACCTCGTCTGACTGTGATATGGAGAATATAATTTTACAAGAGGTACCTTTAGAGGAACAGGAGCATTTCGTGGATTTACAATGCTTCGTAAGCGATGTTGACGCTAAGACGCTGCTGGCTGCTGCGAACCGTCTGGTCAAAGAGAAACCGGACGTTAGATGGCCGGAACTGAACTGGTTTATTGATTTCGTAAAACAAGTCCTAGACTTAGCGACAACTTCAAAAGTTTTAAGTCATCAAACGATGTATGAAAAATTCATTTCATTATTTTCGAGTTATGTGGAGAATTTTGGTGTGAGCTTTACTACCCACATCCTGAACCCGATATTCGTCGATATTATAATGGATCTCGAGCAGAAAATGGAGAAACTTCACACTGTAAATGTCGAAAGCTCTGTGGTGGTCGGTATTTATTCAGTCACAGTGTTGCCAGCGATTGGGAATTATTCACAACACGGAGAATTTTTACAGAAATGGGTAATGTACTGTAGTCTTCGCAGTTTCCCGCCTAAAGTATTATCCATTCCAATGAAGTGGGTTTGCGAACATCGGCCAGACACACTTGATTTTTATTTACAACACCTCAGAGAGTTTGCGGCCAGCAGTTGTGAGAGCGCAAGCGGCAGTTCAGTCCGCGTCTACATCTCCAATATTATCACAGAGTTGTTGAACACAACAGAAATAAACGAGAAATGTATAGAACGACAAGTCCTGCCGGCTGTCGTAGCTCTCTTGCATGACGATGATGTGTCGGTCCGCGAAGTGGCACTCACCTCATGGGGCAGTATGACTCGCACCTGTTTGATCCGAGGGCTGGCGTGCGAAGCGCAGTGCTGGGCCGCGGCCGACGACCTGCTGTGCGGTACACACGTGCTCTCCACGAAGGAAGGGCTGCGAGCGGCCGAGGCACTAACTCTATTGGTGTTGCCCACTGCCGATAGCCACGCCGTCTCAGAGAAAGCGGTGTCGCTACTGTGCGCCCTGTGCGAGCGCGGCCCCGCATGCGCCGAGCGCGTGGCAGCCCTGACGCCGGGCCTGCAGCTGGCGGTGCACCACTGCGGGCGCCACCCGGCCATACTGCCCGCATTAAGAAAATTGGAGGAGGAGGTTCAGTCGCCCGCGCTGTCGCAGCACCGGGCGGCCGTGGAGAGCCTGGTGCAGACGGCGGCCGGCGCGCCCGCCGAGCCCGCGCGCGACCCCCCGCGCCCCGCCAACCTGCTCGCCGCGCAGGAGGTCGGCAGGAGGGTCACGCAGATGTTCCACCAATCCAAGACCAACATCAATTTACAGAATATATTTAAAAAGAAACCGTAG
Protein
MNPYKETEESSFDTPHLSYEDIATKLLKDNFFLTALELHTELVESGKELPQLREFFSNPGNFEQHVSRASELNSIHRTPSQATLDSLDTARYSEDGGGDRGGSGGDVAVLEFELRKARETINSLRANLTQFADGSSTLDKSSQKICDQRSLKPHEKRALNFLINEYLLMQDYKLTSITFSDENPDQEFEDWDDVGLNMPRPAGIVSLFRGESVLSVPKVDSSTETNWQQIDAGCQTELDERSICVSCQDSIDHDNDWAAELLIQAEEIEMLKQKAMALEMEKQNFQKLYDTAIVSLNSLASPAIESKALDLNTKDEKMIEAEQAIDTTPITCTASENHYGSGNSTHSATPEQFEMIESEKNSTITRKEGSTLSSFEHTVLGENCNRDSPLRRGSVTTLDEALSINDGGEWTRVQYEYNSIETSSKEMWIDSGLPNSLKVSILNWCCETLNLNESFSNDLLVELINNDKPITLPGFLTLAADTMPRIIPHTLVARRFEAAALLAASVALLPTGDPRRARTLHSLLTLVKKPEPSDTRSICEAACYIVKWGGSGEVLSSVAELLASRSAERRILASQICVAIAPYVPIELCTSLLLSLVVLMCESNEIEIRSLALKSACLICPVAEHKYAQLENIMFGFLKDPIEKHVKDTVSIFGPILARSALMAGKFCTDLCTRIMRNAAKESSDNEWKNAALYLDLMKSLVLAKLAYVINVQIIKDATSSDCDMENIILQEVPLEEQEHFVDLQCFVSDVDAKTLLAAANRLVKEKPDVRWPELNWFIDFVKQVLDLATTSKVLSHQTMYEKFISLFSSYVENFGVSFTTHILNPIFVDIIMDLEQKMEKLHTVNVESSVVVGIYSVTVLPAIGNYSQHGEFLQKWVMYCSLRSFPPKVLSIPMKWVCEHRPDTLDFYLQHLREFAASSCESASGSSVRVYISNIITELLNTTEINEKCIERQVLPAVVALLHDDDVSVREVALTSWGSMTRTCLIRGLACEAQCWAAADDLLCGTHVLSTKEGLRAAEALTLLVLPTADSHAVSEKAVSLLCALCERGPACAERVAALTPGLQLAVHHCGRHPAILPALRKLEEEVQSPALSQHRAAVESLVQTAAGAPAEPARDPPRPANLLAAQEVGRRVTQMFHQSKTNINLQNIFKKKP
Summary
Uniprot
Pubmed
EMBL
ODYU01008647
SOQ52412.1
AGBW02013617
OWR43141.1
KQ461190
KPJ07249.1
+ More
KK853239 KDR09486.1 KQ971321 EFA00181.1 NEVH01003760 PNF40001.1 KQ761738 OAD57133.1 GL888237 EGI64231.1 KQ435732 KOX77453.1 QOIP01000008 RLU19543.1 KK107109 EZA59311.1 ADTU01002880 KQ979074 KYN22771.1 KQ976565 KYM80526.1 KZ288230 PBC31584.1 LBMM01000342 LBMM01000325 KMR00159.1 KMR01327.1 GEZM01073871 JAV64761.1 GL448096 EFN85414.1 KQ414669 KOC64510.1 KQ981215 KYN44531.1 GL439118 EFN67746.1 NNAY01000544 OXU27774.1 AAZX01001307
KK853239 KDR09486.1 KQ971321 EFA00181.1 NEVH01003760 PNF40001.1 KQ761738 OAD57133.1 GL888237 EGI64231.1 KQ435732 KOX77453.1 QOIP01000008 RLU19543.1 KK107109 EZA59311.1 ADTU01002880 KQ979074 KYN22771.1 KQ976565 KYM80526.1 KZ288230 PBC31584.1 LBMM01000342 LBMM01000325 KMR00159.1 KMR01327.1 GEZM01073871 JAV64761.1 GL448096 EFN85414.1 KQ414669 KOC64510.1 KQ981215 KYN44531.1 GL439118 EFN67746.1 NNAY01000544 OXU27774.1 AAZX01001307
Proteomes
Pfam
PF02985 HEAT
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
Ontologies
KEGG
GO
PANTHER
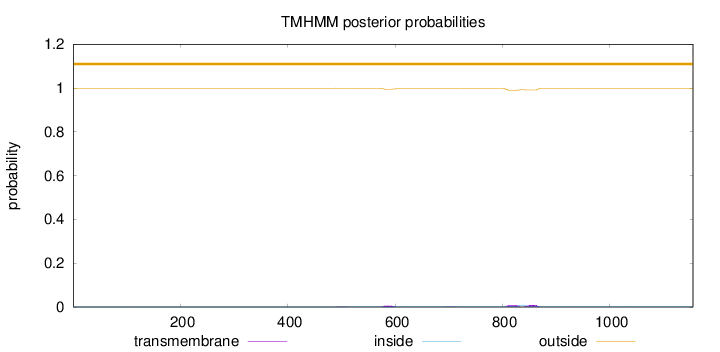
Topology
Length:
1155
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.50129
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00049
outside
1 - 1155
Population Genetic Test Statistics
Pi
266.031945
Theta
186.30439
Tajima's D
1.551926
CLR
0.512259
CSRT
0.797310134493275
Interpretation
Uncertain
