Pre Gene Modal
BGIBMGA011651
Annotation
PREDICTED:_anamorsin_homolog_[Plutella_xylostella]
Full name
Anamorsin homolog
+ More
Anamorsin
Anamorsin
Alternative Name
Fe-S cluster assembly protein DRE2 homolog
Cytokine-induced apoptosis inhibitor 1
Cytokine-induced apoptosis inhibitor 1
Location in the cell
Nuclear Reliability : 2.65
Sequence
CDS
ATGGACAACTTGAAAAACGGAGACAATGTTCTTATTATTTGGAAGAACGACAGTCAAGGAAACATTACGAGCCTAATAGAAGACGTAAAAAAAGTAATTCAAAGTGATACTGTGGTTCTGGAAAACTCCGACATGATCACTGAAGGTTCCCGTGAGCATTCATCATTTGATGTTGTTCTCTCAAACTGGTTACCTCCCCACAGTGTCTCACATTCTGATAAACTATTGGCAATAATCATTAAATTACTCAAACCAAATGGTAAACTTATACTCAAAGATCTCAACAAATTGACCAGTACACTTAAATTAAATGGTTTTGTAAATGTCACTGAATTGGAGAATAACAGATATGTTGCGGAGAAACCAAAGTTTGAGGTTGGTTCAAAGTCATCTCTGAAATTAAATGGAAAAGCCACAGTCTGGAAGTTGGACAGCACAGTTGAGGAAGCTTGGGCCAGCACTAACAACGATGATATTATTGATGAGAACGAACTATTGGATGAAAATGACCTTAAAAAGCCAGATAAAGAAGCTTTGAGTGTTTGTGCAACAACAGGCAAGAGAAAAGCGTGCGTGGACTGCACCTGTGGCCTCGCCGAGGAACTAAGGGGCGAGACAAAAGAAACTCCAAAATCGTCGTGTGGAAGTTGTTACTTGGGCGATGCGTTCCGATGCGCGACCTGCCCGTACCTCGGCATGCCAGCCTTCAAGCCCGGCGAGAAAGTGATGTTAGATCTCAAGGCCGATATATGA
Protein
MDNLKNGDNVLIIWKNDSQGNITSLIEDVKKVIQSDTVVLENSDMITEGSREHSSFDVVLSNWLPPHSVSHSDKLLAIIIKLLKPNGKLILKDLNKLTSTLKLNGFVNVTELENNRYVAEKPKFEVGSKSSLKLNGKATVWKLDSTVEEAWASTNNDDIIDENELLDENDLKKPDKEALSVCATTGKRKACVDCTCGLAEELRGETKETPKSSCGSCYLGDAFRCATCPYLGMPAFKPGEKVMLDLKADI
Summary
Description
Component of the cytosolic iron-sulfur (Fe-S) protein assembly (CIA) machinery. Required for the maturation of extramitochondrial Fe-S proteins. Part of an electron transfer chain functioning in an early step of cytosolic Fe-S biogenesis, facilitating the de novo assembly of a [4Fe-4S] cluster on the cytosolic Fe-S scaffold complex. Electrons are transferred from NADPH via a FAD- and FMN-containing diflavin oxidoreductase. Together with the diflavin oxidoreductase, also required for the assembly of the diferric tyrosyl radical cofactor of ribonucleotide reductase (RNR), probably by providing electrons for reduction during radical cofactor maturation in the catalytic small subunit.
Component of the cytosolic iron-sulfur (Fe-S) protein assembly (CIA) machinery required for the maturation of extramitochondrial Fe-S proteins. Part of an electron transfer chain functioning in an early step of cytosolic Fe-S biogenesis, facilitating the de novo assembly of a [4Fe-4S] cluster on the scaffold complex NUBP1-NUBP2. Electrons are transferred to CIAPIN1 from NADPH via the FAD- and FMN-containing protein NDOR1. NDOR1-CIAPIN1 are also required for the assembly of the diferric tyrosyl radical cofactor of ribonucleotide reductase (RNR), probably by providing electrons for reduction during radical cofactor maturation in the catalytic small subunit. Has anti-apoptotic effects in the cell. Involved in negative control of cell death upon cytokine withdrawal. Promotes development of hematopoietic cells.
Component of the cytosolic iron-sulfur (Fe-S) protein assembly (CIA) machinery required for the maturation of extramitochondrial Fe-S proteins. Part of an electron transfer chain functioning in an early step of cytosolic Fe-S biogenesis, facilitating the de novo assembly of a [4Fe-4S] cluster on the scaffold complex NUBP1-NUBP2. Electrons are transferred to CIAPIN1 from NADPH via the FAD- and FMN-containing protein NDOR1. NDOR1-CIAPIN1 are also required for the assembly of the diferric tyrosyl radical cofactor of ribonucleotide reductase (RNR), probably by providing electrons for reduction during radical cofactor maturation in the catalytic small subunit. Has anti-apoptotic effects in the cell. Involved in negative control of cell death upon cytokine withdrawal. Promotes development of hematopoietic cells.
Cofactor
[2Fe-2S] cluster
[4Fe-4S] cluster
[4Fe-4S] cluster
Subunit
Monomer.
Monomer. Interacts with NDOR1. Interacts with CHCHD4.
Monomer. Interacts with NDOR1. Interacts with CHCHD4.
Similarity
Belongs to the anamorsin family.
Keywords
2Fe-2S
4Fe-4S
Complete proteome
Cytoplasm
Iron
Iron-sulfur
Metal-binding
Mitochondrion
Reference proteome
Feature
chain Anamorsin homolog
Uniprot
H9JQ45
A0A2W1BP47
A0A2A4JAH4
A0A2H1WPT8
I4DLD1
I4DMN8
+ More
A0A3S2L842 A0A194QUM9 S4PBA6 A0A336MID5 A0A336L2I2 A0A182H5V0 A0A023ENN9 Q17L24 U5EZP7 A0A1L8DJE0 A0A182JL34 R4G435 A0A0P4VK93 A0A1L8DJ86 A0A224XTC9 A0A2J7PDG1 A0A182VUA7 A0A084VYN7 A0A1Q3FHC1 A0A182Y932 A0A182P675 E0VVV1 B0WQ75 A0A164U5S6 A0A182M6Q8 A0A2M3ZAR1 A0A2M3ZIR8 A0A182JSP8 A0A182QA31 A0A182NGL8 W5JDU4 A0A182RUQ0 A0A182F5S4 A0A182VLU3 Q7PWQ6 A0A0P6BHU6 A0A2M4AL90 T1GFK3 A0A182XDC5 A0A182LMX8 A0A182UKD8 A0A182I776 A0A2M4BY03 A0A2M4BY08 A0A2M4ALW0 A0A067RKU9 A0A0T6B1T1 A0A2M4ANS4 A0A1J1HRV3 A0A0K8TA45 A0A0A9Z9I8 A0A3B4DTP7 A0A2D0RE45 A0A1B0CE17 E9G038 E3TGJ0 A0A212ENS5 A0A091HPD0 A0A1B6HMY8 A0A2M4AMY9 A0A093QT01 A0A2D0RF89 A0A1V9XF13 A0A093DLM5 A0A0L7R1C9 K7IZS4 A0A0L0C6D9 A0A1A7XQD0 D6WCU7 W5L046 A0A3B1JES3 A0A093JDV6 A0A232FFP7 R0JU51 A0A093HBM5 A0A091MPX2 A0A0P6I6S5 A0A1B6G9N3 A0A093C8X6 A0A091WN69 A0A091FLL3 A0A315VZG0 K1PUR4 A0A093NG25 A0A087QN74 A0A3Q3IMV3 A0A1V4K0U4 B4NYT3 B4HXL7 M4ANG0 A0A091R1B5 A0A2I4D9Y4 A0A087VLS0 A0A0L7LGC4
A0A3S2L842 A0A194QUM9 S4PBA6 A0A336MID5 A0A336L2I2 A0A182H5V0 A0A023ENN9 Q17L24 U5EZP7 A0A1L8DJE0 A0A182JL34 R4G435 A0A0P4VK93 A0A1L8DJ86 A0A224XTC9 A0A2J7PDG1 A0A182VUA7 A0A084VYN7 A0A1Q3FHC1 A0A182Y932 A0A182P675 E0VVV1 B0WQ75 A0A164U5S6 A0A182M6Q8 A0A2M3ZAR1 A0A2M3ZIR8 A0A182JSP8 A0A182QA31 A0A182NGL8 W5JDU4 A0A182RUQ0 A0A182F5S4 A0A182VLU3 Q7PWQ6 A0A0P6BHU6 A0A2M4AL90 T1GFK3 A0A182XDC5 A0A182LMX8 A0A182UKD8 A0A182I776 A0A2M4BY03 A0A2M4BY08 A0A2M4ALW0 A0A067RKU9 A0A0T6B1T1 A0A2M4ANS4 A0A1J1HRV3 A0A0K8TA45 A0A0A9Z9I8 A0A3B4DTP7 A0A2D0RE45 A0A1B0CE17 E9G038 E3TGJ0 A0A212ENS5 A0A091HPD0 A0A1B6HMY8 A0A2M4AMY9 A0A093QT01 A0A2D0RF89 A0A1V9XF13 A0A093DLM5 A0A0L7R1C9 K7IZS4 A0A0L0C6D9 A0A1A7XQD0 D6WCU7 W5L046 A0A3B1JES3 A0A093JDV6 A0A232FFP7 R0JU51 A0A093HBM5 A0A091MPX2 A0A0P6I6S5 A0A1B6G9N3 A0A093C8X6 A0A091WN69 A0A091FLL3 A0A315VZG0 K1PUR4 A0A093NG25 A0A087QN74 A0A3Q3IMV3 A0A1V4K0U4 B4NYT3 B4HXL7 M4ANG0 A0A091R1B5 A0A2I4D9Y4 A0A087VLS0 A0A0L7LGC4
Pubmed
19121390
28756777
22651552
26354079
23622113
26483478
+ More
24945155 17510324 27129103 24438588 25244985 20566863 20920257 23761445 12364791 20966253 24845553 25401762 21292972 20634964 22118469 28327890 20075255 26108605 18362917 19820115 25329095 28648823 29703783 22992520 17994087 23542700 26227816
24945155 17510324 27129103 24438588 25244985 20566863 20920257 23761445 12364791 20966253 24845553 25401762 21292972 20634964 22118469 28327890 20075255 26108605 18362917 19820115 25329095 28648823 29703783 22992520 17994087 23542700 26227816
EMBL
BABH01012138
BABH01012139
KZ149971
PZC76051.1
NWSH01002125
PCG69085.1
+ More
ODYU01010147 SOQ55048.1 AK402099 KQ459590 BAM18721.1 KPI97282.1 AK402556 BAM19178.1 RSAL01000094 RVE47875.1 KQ461190 KPJ07261.1 GAIX01006007 JAA86553.1 UFQT01001355 SSX30182.1 UFQS01000874 UFQT01000874 UFQT01004235 SSX07420.1 SSX27762.1 SSX35586.1 JXUM01026690 KQ560764 KXJ80974.1 GAPW01003554 JAC10044.1 CH477219 GANO01000396 JAB59475.1 GFDF01007543 JAV06541.1 ACPB03012557 GAHY01001136 JAA76374.1 GDKW01001124 JAI55471.1 GFDF01007545 JAV06539.1 GFTR01004644 JAW11782.1 NEVH01026387 PNF14370.1 ATLV01018382 KE525231 KFB43081.1 GFDL01008097 JAV26948.1 DS235815 EEB17507.1 DS232036 LRGB01001581 KZS11086.1 AXCM01000336 GGFM01004868 MBW25619.1 GGFM01007648 MBW28399.1 AXCN02002086 ADMH02001840 ETN60954.1 AAAB01008984 GDIP01015677 JAM88038.1 GGFK01008234 MBW41555.1 CAQQ02395050 APCN01003368 GGFJ01008751 MBW57892.1 GGFJ01008752 MBW57893.1 GGFK01008440 MBW41761.1 KK852567 KDR21215.1 LJIG01016192 KRT81340.1 GGFK01009061 MBW42382.1 CVRI01000020 CRK90707.1 GBRD01003540 JAG62281.1 GBHO01001662 JAG41942.1 AJWK01008508 GL732528 EFX87463.1 GU589476 ADO29426.1 AGBW02013617 OWR43129.1 KL519666 KFO89178.1 GECU01031680 JAS76026.1 GGFK01008832 MBW42153.1 KL431080 KFW91641.1 MNPL01013003 OQR71948.1 KN127047 KFU94711.1 KQ414668 KOC64647.1 JRES01000940 KNC26989.1 HADW01018649 HADX01011977 SBP20049.1 KQ971311 EEZ99063.1 KK575366 KFW10037.1 NNAY01000319 OXU29259.1 KB743139 EOB01001.1 KL205902 KFV76367.1 KK834474 KFP79010.1 GDIQ01009701 JAN85036.1 GECZ01022160 GECZ01010627 JAS47609.1 JAS59142.1 KL455283 KFV08747.1 KL411055 KFR02951.1 KL447260 KFO71365.1 NHOQ01001396 PWA24856.1 JH815838 EKC25468.1 KL224535 KFW60965.1 KL225750 KFM02678.1 LSYS01005191 OPJ78024.1 CM000157 CH480818 KK711337 KFQ33565.1 KL497913 KFO13562.1 JTDY01001239 KOB74439.1
ODYU01010147 SOQ55048.1 AK402099 KQ459590 BAM18721.1 KPI97282.1 AK402556 BAM19178.1 RSAL01000094 RVE47875.1 KQ461190 KPJ07261.1 GAIX01006007 JAA86553.1 UFQT01001355 SSX30182.1 UFQS01000874 UFQT01000874 UFQT01004235 SSX07420.1 SSX27762.1 SSX35586.1 JXUM01026690 KQ560764 KXJ80974.1 GAPW01003554 JAC10044.1 CH477219 GANO01000396 JAB59475.1 GFDF01007543 JAV06541.1 ACPB03012557 GAHY01001136 JAA76374.1 GDKW01001124 JAI55471.1 GFDF01007545 JAV06539.1 GFTR01004644 JAW11782.1 NEVH01026387 PNF14370.1 ATLV01018382 KE525231 KFB43081.1 GFDL01008097 JAV26948.1 DS235815 EEB17507.1 DS232036 LRGB01001581 KZS11086.1 AXCM01000336 GGFM01004868 MBW25619.1 GGFM01007648 MBW28399.1 AXCN02002086 ADMH02001840 ETN60954.1 AAAB01008984 GDIP01015677 JAM88038.1 GGFK01008234 MBW41555.1 CAQQ02395050 APCN01003368 GGFJ01008751 MBW57892.1 GGFJ01008752 MBW57893.1 GGFK01008440 MBW41761.1 KK852567 KDR21215.1 LJIG01016192 KRT81340.1 GGFK01009061 MBW42382.1 CVRI01000020 CRK90707.1 GBRD01003540 JAG62281.1 GBHO01001662 JAG41942.1 AJWK01008508 GL732528 EFX87463.1 GU589476 ADO29426.1 AGBW02013617 OWR43129.1 KL519666 KFO89178.1 GECU01031680 JAS76026.1 GGFK01008832 MBW42153.1 KL431080 KFW91641.1 MNPL01013003 OQR71948.1 KN127047 KFU94711.1 KQ414668 KOC64647.1 JRES01000940 KNC26989.1 HADW01018649 HADX01011977 SBP20049.1 KQ971311 EEZ99063.1 KK575366 KFW10037.1 NNAY01000319 OXU29259.1 KB743139 EOB01001.1 KL205902 KFV76367.1 KK834474 KFP79010.1 GDIQ01009701 JAN85036.1 GECZ01022160 GECZ01010627 JAS47609.1 JAS59142.1 KL455283 KFV08747.1 KL411055 KFR02951.1 KL447260 KFO71365.1 NHOQ01001396 PWA24856.1 JH815838 EKC25468.1 KL224535 KFW60965.1 KL225750 KFM02678.1 LSYS01005191 OPJ78024.1 CM000157 CH480818 KK711337 KFQ33565.1 KL497913 KFO13562.1 JTDY01001239 KOB74439.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000053240
UP000069940
+ More
UP000249989 UP000008820 UP000075880 UP000015103 UP000235965 UP000075920 UP000030765 UP000076408 UP000075885 UP000009046 UP000002320 UP000076858 UP000075883 UP000075881 UP000075886 UP000075884 UP000000673 UP000075900 UP000069272 UP000075903 UP000007062 UP000015102 UP000076407 UP000075882 UP000075902 UP000075840 UP000027135 UP000183832 UP000261440 UP000221080 UP000092461 UP000000305 UP000007151 UP000192247 UP000053825 UP000002358 UP000037069 UP000007266 UP000018467 UP000215335 UP000053584 UP000053283 UP000053760 UP000005408 UP000054081 UP000053286 UP000261600 UP000190648 UP000002282 UP000001292 UP000002852 UP000192220 UP000037510
UP000249989 UP000008820 UP000075880 UP000015103 UP000235965 UP000075920 UP000030765 UP000076408 UP000075885 UP000009046 UP000002320 UP000076858 UP000075883 UP000075881 UP000075886 UP000075884 UP000000673 UP000075900 UP000069272 UP000075903 UP000007062 UP000015102 UP000076407 UP000075882 UP000075902 UP000075840 UP000027135 UP000183832 UP000261440 UP000221080 UP000092461 UP000000305 UP000007151 UP000192247 UP000053825 UP000002358 UP000037069 UP000007266 UP000018467 UP000215335 UP000053584 UP000053283 UP000053760 UP000005408 UP000054081 UP000053286 UP000261600 UP000190648 UP000002282 UP000001292 UP000002852 UP000192220 UP000037510
Interpro
Gene 3D
ProteinModelPortal
H9JQ45
A0A2W1BP47
A0A2A4JAH4
A0A2H1WPT8
I4DLD1
I4DMN8
+ More
A0A3S2L842 A0A194QUM9 S4PBA6 A0A336MID5 A0A336L2I2 A0A182H5V0 A0A023ENN9 Q17L24 U5EZP7 A0A1L8DJE0 A0A182JL34 R4G435 A0A0P4VK93 A0A1L8DJ86 A0A224XTC9 A0A2J7PDG1 A0A182VUA7 A0A084VYN7 A0A1Q3FHC1 A0A182Y932 A0A182P675 E0VVV1 B0WQ75 A0A164U5S6 A0A182M6Q8 A0A2M3ZAR1 A0A2M3ZIR8 A0A182JSP8 A0A182QA31 A0A182NGL8 W5JDU4 A0A182RUQ0 A0A182F5S4 A0A182VLU3 Q7PWQ6 A0A0P6BHU6 A0A2M4AL90 T1GFK3 A0A182XDC5 A0A182LMX8 A0A182UKD8 A0A182I776 A0A2M4BY03 A0A2M4BY08 A0A2M4ALW0 A0A067RKU9 A0A0T6B1T1 A0A2M4ANS4 A0A1J1HRV3 A0A0K8TA45 A0A0A9Z9I8 A0A3B4DTP7 A0A2D0RE45 A0A1B0CE17 E9G038 E3TGJ0 A0A212ENS5 A0A091HPD0 A0A1B6HMY8 A0A2M4AMY9 A0A093QT01 A0A2D0RF89 A0A1V9XF13 A0A093DLM5 A0A0L7R1C9 K7IZS4 A0A0L0C6D9 A0A1A7XQD0 D6WCU7 W5L046 A0A3B1JES3 A0A093JDV6 A0A232FFP7 R0JU51 A0A093HBM5 A0A091MPX2 A0A0P6I6S5 A0A1B6G9N3 A0A093C8X6 A0A091WN69 A0A091FLL3 A0A315VZG0 K1PUR4 A0A093NG25 A0A087QN74 A0A3Q3IMV3 A0A1V4K0U4 B4NYT3 B4HXL7 M4ANG0 A0A091R1B5 A0A2I4D9Y4 A0A087VLS0 A0A0L7LGC4
A0A3S2L842 A0A194QUM9 S4PBA6 A0A336MID5 A0A336L2I2 A0A182H5V0 A0A023ENN9 Q17L24 U5EZP7 A0A1L8DJE0 A0A182JL34 R4G435 A0A0P4VK93 A0A1L8DJ86 A0A224XTC9 A0A2J7PDG1 A0A182VUA7 A0A084VYN7 A0A1Q3FHC1 A0A182Y932 A0A182P675 E0VVV1 B0WQ75 A0A164U5S6 A0A182M6Q8 A0A2M3ZAR1 A0A2M3ZIR8 A0A182JSP8 A0A182QA31 A0A182NGL8 W5JDU4 A0A182RUQ0 A0A182F5S4 A0A182VLU3 Q7PWQ6 A0A0P6BHU6 A0A2M4AL90 T1GFK3 A0A182XDC5 A0A182LMX8 A0A182UKD8 A0A182I776 A0A2M4BY03 A0A2M4BY08 A0A2M4ALW0 A0A067RKU9 A0A0T6B1T1 A0A2M4ANS4 A0A1J1HRV3 A0A0K8TA45 A0A0A9Z9I8 A0A3B4DTP7 A0A2D0RE45 A0A1B0CE17 E9G038 E3TGJ0 A0A212ENS5 A0A091HPD0 A0A1B6HMY8 A0A2M4AMY9 A0A093QT01 A0A2D0RF89 A0A1V9XF13 A0A093DLM5 A0A0L7R1C9 K7IZS4 A0A0L0C6D9 A0A1A7XQD0 D6WCU7 W5L046 A0A3B1JES3 A0A093JDV6 A0A232FFP7 R0JU51 A0A093HBM5 A0A091MPX2 A0A0P6I6S5 A0A1B6G9N3 A0A093C8X6 A0A091WN69 A0A091FLL3 A0A315VZG0 K1PUR4 A0A093NG25 A0A087QN74 A0A3Q3IMV3 A0A1V4K0U4 B4NYT3 B4HXL7 M4ANG0 A0A091R1B5 A0A2I4D9Y4 A0A087VLS0 A0A0L7LGC4
PDB
2YUI
E-value=0.00100534,
Score=98
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Mitochondrion intermembrane space
Nucleus
Mitochondrion intermembrane space
Nucleus
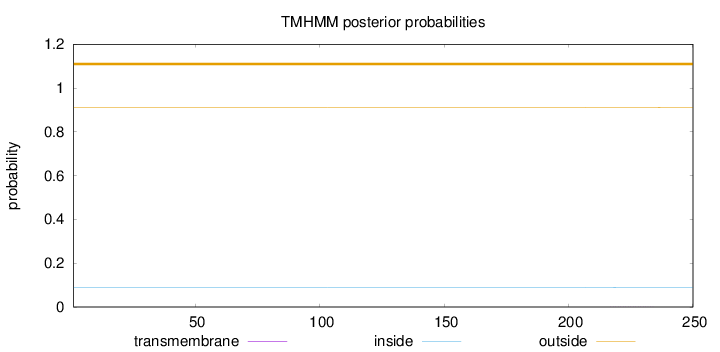
Length:
250
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00337
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08975
outside
1 - 250
Population Genetic Test Statistics
Pi
200.597813
Theta
173.09774
Tajima's D
0.648275
CLR
0
CSRT
0.56177191140443
Interpretation
Uncertain
