Gene
KWMTBOMO06551
Annotation
PREDICTED:_dual_specificity_protein_phosphatase_22_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.464
Sequence
CDS
ATGGGAAATGGAATGAACAAGGTACTCCCCGGCCTATACGTGGGCAACTACAGAGACTCTAAGGACACCGTACAGCTAGAGAAATACAAGATAACACACATACTATCCATACACGATGCCGCAAGAAGACTACACTCTGACAAGCACTACCTCTGCATAATGGCGTCTGATTCACCAGACCAGAACTTGACACAGTACTTCAGTTTATGTAACGACTTCATACACGCCGCGAGACTCAGAGAAGGCAACGTCCTTATACATTGTTTGGCGGGAATGTCTCGGAGCGTGACAGTTGCCGTTGCGTACATAATGTCGGTGACACCCCTAACATGGCGAGAGGCATTGAAAGTTGTCAGAGCGGGTCGAGCTGTTGCCAACCCTAATCTCGGCTTCCAGAGGCAACTCCAAGACTTCGAAACTTACAAACTAGTTGAGGAGAGGAGACGTTTAAAAGAACGGTATCCGTCACTGGCTCTAGCCGAGAAGGATTTATCGGAATGCCGCATCATGCTCGATTCCTACCAGACGATGCTGAGCGAGAGAACCATTTGCGAAGGAAAGTGCGCCATGGGCCGGCAGTGTCCAACTGGTAAGGCACAAAGTATGAATTTGGAATATTTACCGCGCGAACGAACACAAATTTCGACGATGGCAAAAGCCTCTAAACCTTTGAACACTTCAAGCTTCCTTCCTTTATGA
Protein
MGNGMNKVLPGLYVGNYRDSKDTVQLEKYKITHILSIHDAARRLHSDKHYLCIMASDSPDQNLTQYFSLCNDFIHAARLREGNVLIHCLAGMSRSVTVAVAYIMSVTPLTWREALKVVRAGRAVANPNLGFQRQLQDFETYKLVEERRRLKERYPSLALAEKDLSECRIMLDSYQTMLSERTICEGKCAMGRQCPTGKAQSMNLEYLPRERTQISTMAKASKPLNTSSFLPL
Summary
Similarity
Belongs to the protein-tyrosine phosphatase family. Non-receptor class dual specificity subfamily.
Uniprot
A0A2A4IZK7
A0A194QP09
A0A1W4W8G9
A0A139WL97
A0A0C9R0F2
A0A139WLH8
+ More
A0A1W4W8F6 D6WHC9 A0A0C9QZX4 K7IXZ8 N6U264 A0A2P8XIA8 A0A3L8DY15 Q17M44 A0A158N9C2 A0A1B6E338 A0A2J7PCW7 A0A1B0C9F7 T1P9R3 A0A1B0FPK4 A0A1A9ZRW6 A0A0Q9XLX8 E0VL47 A0A0A1WGE2 A0A0K8VYC3 A0A026WC91 Q8IQK1 A0A1I8MZT9 A0A1A9WQ28 W8AZD7 A0A1A9YIE8 A0A1A9UG79 B4J2Z7 A0A154NYU4 A0A1I8Q8J0 A0A0Q9WTA2 A0A1B0AQX4 A0A0L0CDE5 B4LFI5 B4PHG2 A0A195FUU5 A0A1W4VKE4 B3NHG2 Q9VU80 A0A0L7R825 A0A182UJF7 A0A182KST7 B4HH71 B4QJF9 A0A1S4HE00 A0A0P4VLC1 A0A232F012 E2B8S2 A0A0M4EHX2 B4MNC5 A0A151X9W6 B3MB37 F4WM09 B4KX60 A0A195BC47 A0A1W4VJ08 A0A023F303 A0A0A1XAU0 A0A0K8VAE1 A0A0R3P6F5 Q29E18 A0A0Q9XDW2 B4H423 A0A182JEY7 W5J591 Q8IQK0 A0A0A9YY25 A0A0Q9WWT4 A0A336LIR5 B0WEK1 A0A182H6A6 A0A1W4V6V8 A0A0M9ACA6 M9PI51 A0A0J9RUX6 A0A0Q5U824 A0A310S9S3 A0A182RGL3 A0A0Q9WK53 A0A0R1DZ98 A0A182HPY6 A0A182XDA3 A0A182FUF5 A0A0P8XWC3 A0A067R3G5 A0A182W9A0 A0A195C660 A0A0P8YJB2 A0A182PBM7 A0A182QDZ6 A0A146L973
A0A1W4W8F6 D6WHC9 A0A0C9QZX4 K7IXZ8 N6U264 A0A2P8XIA8 A0A3L8DY15 Q17M44 A0A158N9C2 A0A1B6E338 A0A2J7PCW7 A0A1B0C9F7 T1P9R3 A0A1B0FPK4 A0A1A9ZRW6 A0A0Q9XLX8 E0VL47 A0A0A1WGE2 A0A0K8VYC3 A0A026WC91 Q8IQK1 A0A1I8MZT9 A0A1A9WQ28 W8AZD7 A0A1A9YIE8 A0A1A9UG79 B4J2Z7 A0A154NYU4 A0A1I8Q8J0 A0A0Q9WTA2 A0A1B0AQX4 A0A0L0CDE5 B4LFI5 B4PHG2 A0A195FUU5 A0A1W4VKE4 B3NHG2 Q9VU80 A0A0L7R825 A0A182UJF7 A0A182KST7 B4HH71 B4QJF9 A0A1S4HE00 A0A0P4VLC1 A0A232F012 E2B8S2 A0A0M4EHX2 B4MNC5 A0A151X9W6 B3MB37 F4WM09 B4KX60 A0A195BC47 A0A1W4VJ08 A0A023F303 A0A0A1XAU0 A0A0K8VAE1 A0A0R3P6F5 Q29E18 A0A0Q9XDW2 B4H423 A0A182JEY7 W5J591 Q8IQK0 A0A0A9YY25 A0A0Q9WWT4 A0A336LIR5 B0WEK1 A0A182H6A6 A0A1W4V6V8 A0A0M9ACA6 M9PI51 A0A0J9RUX6 A0A0Q5U824 A0A310S9S3 A0A182RGL3 A0A0Q9WK53 A0A0R1DZ98 A0A182HPY6 A0A182XDA3 A0A182FUF5 A0A0P8XWC3 A0A067R3G5 A0A182W9A0 A0A195C660 A0A0P8YJB2 A0A182PBM7 A0A182QDZ6 A0A146L973
Pubmed
26354079
18362917
19820115
20075255
23537049
29403074
+ More
30249741 17510324 21347285 17994087 20566863 25830018 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 24495485 26108605 17550304 20966253 22936249 12364791 27129103 28648823 20798317 21719571 25474469 15632085 20920257 23761445 25401762 26483478 24845553 26823975
30249741 17510324 21347285 17994087 20566863 25830018 24508170 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 24495485 26108605 17550304 20966253 22936249 12364791 27129103 28648823 20798317 21719571 25474469 15632085 20920257 23761445 25401762 26483478 24845553 26823975
EMBL
NWSH01004467
PCG65069.1
KQ461190
KPJ07263.1
KQ971321
KYB28676.1
+ More
GBYB01006417 JAG76184.1 KYB28677.1 EFA00642.2 GBYB01006242 JAG76009.1 APGK01042703 APGK01042704 APGK01042705 KB741007 KB631811 ENN75625.1 ERL86345.1 PYGN01002038 PSN31730.1 QOIP01000002 RLU25350.1 CH477208 EAT47801.1 ADTU01001372 GEDC01004967 JAS32331.1 NEVH01026401 PNF14174.1 AJWK01002279 AJWK01002280 KA645486 AFP60115.1 CCAG010022945 CH933809 KRG06746.1 DS235269 EEB14103.1 GBXI01016819 JAC97472.1 GDHF01008478 JAI43836.1 KK107321 EZA52654.1 AE014296 BT014928 AAN11825.1 AAN11826.1 AAT47779.1 GAMC01016332 JAB90223.1 CH916366 EDV97167.1 KQ434777 KZC04188.1 CH940647 KRF84833.1 JXJN01002101 JRES01000558 KNC30242.1 EDW70303.1 CM000159 EDW94423.1 KQ981264 KYN44062.1 CH954178 EDV51688.1 BT126363 AAF49810.2 AEC46879.1 KQ414637 KOC66983.1 CH480815 EDW41396.1 CM000363 CM002912 EDX10368.1 KMY99406.1 AAAB01008986 GDKW01003173 JAI53422.1 NNAY01001442 OXU23965.1 GL446371 EFN87917.1 CP012525 ALC44252.1 CH963847 EDW72634.1 KQ982353 KYQ57171.1 CH902618 EDV41338.1 GL888217 EGI64653.1 EDW19703.2 KQ976519 KYM82123.1 GBBI01003109 JAC15603.1 GBXI01006055 JAD08237.1 GDHF01026835 GDHF01016481 GDHF01016385 JAI25479.1 JAI35833.1 JAI35929.1 CH379070 KRT08737.1 EAL30245.3 KRG06747.1 CH479208 EDW31138.1 ADMH02002188 ETN57909.1 AAN11827.1 GBHO01007606 JAG35998.1 KRF84832.1 UFQT01000022 SSX18014.1 DS231910 EDS45717.1 JXUM01026849 JXUM01026850 KQ560769 KXJ80956.1 KQ435694 KOX80916.1 AGB94496.1 KMY99407.1 KQS43892.1 KQ766607 OAD53618.1 KRF84834.1 KRK01806.1 APCN01003284 KPU79063.1 KK852729 KDR17551.1 KQ978220 KYM96337.1 KPU79064.1 AXCN02001826 GDHC01014853 JAQ03776.1
GBYB01006417 JAG76184.1 KYB28677.1 EFA00642.2 GBYB01006242 JAG76009.1 APGK01042703 APGK01042704 APGK01042705 KB741007 KB631811 ENN75625.1 ERL86345.1 PYGN01002038 PSN31730.1 QOIP01000002 RLU25350.1 CH477208 EAT47801.1 ADTU01001372 GEDC01004967 JAS32331.1 NEVH01026401 PNF14174.1 AJWK01002279 AJWK01002280 KA645486 AFP60115.1 CCAG010022945 CH933809 KRG06746.1 DS235269 EEB14103.1 GBXI01016819 JAC97472.1 GDHF01008478 JAI43836.1 KK107321 EZA52654.1 AE014296 BT014928 AAN11825.1 AAN11826.1 AAT47779.1 GAMC01016332 JAB90223.1 CH916366 EDV97167.1 KQ434777 KZC04188.1 CH940647 KRF84833.1 JXJN01002101 JRES01000558 KNC30242.1 EDW70303.1 CM000159 EDW94423.1 KQ981264 KYN44062.1 CH954178 EDV51688.1 BT126363 AAF49810.2 AEC46879.1 KQ414637 KOC66983.1 CH480815 EDW41396.1 CM000363 CM002912 EDX10368.1 KMY99406.1 AAAB01008986 GDKW01003173 JAI53422.1 NNAY01001442 OXU23965.1 GL446371 EFN87917.1 CP012525 ALC44252.1 CH963847 EDW72634.1 KQ982353 KYQ57171.1 CH902618 EDV41338.1 GL888217 EGI64653.1 EDW19703.2 KQ976519 KYM82123.1 GBBI01003109 JAC15603.1 GBXI01006055 JAD08237.1 GDHF01026835 GDHF01016481 GDHF01016385 JAI25479.1 JAI35833.1 JAI35929.1 CH379070 KRT08737.1 EAL30245.3 KRG06747.1 CH479208 EDW31138.1 ADMH02002188 ETN57909.1 AAN11827.1 GBHO01007606 JAG35998.1 KRF84832.1 UFQT01000022 SSX18014.1 DS231910 EDS45717.1 JXUM01026849 JXUM01026850 KQ560769 KXJ80956.1 KQ435694 KOX80916.1 AGB94496.1 KMY99407.1 KQS43892.1 KQ766607 OAD53618.1 KRF84834.1 KRK01806.1 APCN01003284 KPU79063.1 KK852729 KDR17551.1 KQ978220 KYM96337.1 KPU79064.1 AXCN02001826 GDHC01014853 JAQ03776.1
Proteomes
UP000218220
UP000053240
UP000192223
UP000007266
UP000002358
UP000019118
+ More
UP000030742 UP000245037 UP000279307 UP000008820 UP000005205 UP000235965 UP000092461 UP000092444 UP000092445 UP000009192 UP000009046 UP000053097 UP000000803 UP000095301 UP000091820 UP000092443 UP000078200 UP000001070 UP000076502 UP000095300 UP000008792 UP000092460 UP000037069 UP000002282 UP000078541 UP000192221 UP000008711 UP000053825 UP000075902 UP000075882 UP000001292 UP000000304 UP000215335 UP000008237 UP000092553 UP000007798 UP000075809 UP000007801 UP000007755 UP000078540 UP000001819 UP000008744 UP000075880 UP000000673 UP000002320 UP000069940 UP000249989 UP000053105 UP000075900 UP000075840 UP000076407 UP000069272 UP000027135 UP000075920 UP000078542 UP000075885 UP000075886
UP000030742 UP000245037 UP000279307 UP000008820 UP000005205 UP000235965 UP000092461 UP000092444 UP000092445 UP000009192 UP000009046 UP000053097 UP000000803 UP000095301 UP000091820 UP000092443 UP000078200 UP000001070 UP000076502 UP000095300 UP000008792 UP000092460 UP000037069 UP000002282 UP000078541 UP000192221 UP000008711 UP000053825 UP000075902 UP000075882 UP000001292 UP000000304 UP000215335 UP000008237 UP000092553 UP000007798 UP000075809 UP000007801 UP000007755 UP000078540 UP000001819 UP000008744 UP000075880 UP000000673 UP000002320 UP000069940 UP000249989 UP000053105 UP000075900 UP000075840 UP000076407 UP000069272 UP000027135 UP000075920 UP000078542 UP000075885 UP000075886
Interpro
IPR000340
Dual-sp_phosphatase_cat-dom
+ More
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000387 TYR_PHOSPHATASE_dom
IPR020417 Atypical_DUSP
IPR029021 Prot-tyrosine_phosphatase-like
IPR003903 UIM_dom
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR000719 Prot_kinase_dom
IPR027417 P-loop_NTPase
IPR007696 DNA_mismatch_repair_MutS_core
IPR000432 DNA_mismatch_repair_MutS_C
IPR036678 MutS_con_dom_sf
IPR036187 DNA_mismatch_repair_MutS_sf
IPR020422 TYR_PHOSPHATASE_DUAL_dom
IPR000387 TYR_PHOSPHATASE_dom
IPR020417 Atypical_DUSP
IPR029021 Prot-tyrosine_phosphatase-like
IPR003903 UIM_dom
IPR011009 Kinase-like_dom_sf
IPR008271 Ser/Thr_kinase_AS
IPR017441 Protein_kinase_ATP_BS
IPR000719 Prot_kinase_dom
IPR027417 P-loop_NTPase
IPR007696 DNA_mismatch_repair_MutS_core
IPR000432 DNA_mismatch_repair_MutS_C
IPR036678 MutS_con_dom_sf
IPR036187 DNA_mismatch_repair_MutS_sf
Gene 3D
CDD
ProteinModelPortal
A0A2A4IZK7
A0A194QP09
A0A1W4W8G9
A0A139WL97
A0A0C9R0F2
A0A139WLH8
+ More
A0A1W4W8F6 D6WHC9 A0A0C9QZX4 K7IXZ8 N6U264 A0A2P8XIA8 A0A3L8DY15 Q17M44 A0A158N9C2 A0A1B6E338 A0A2J7PCW7 A0A1B0C9F7 T1P9R3 A0A1B0FPK4 A0A1A9ZRW6 A0A0Q9XLX8 E0VL47 A0A0A1WGE2 A0A0K8VYC3 A0A026WC91 Q8IQK1 A0A1I8MZT9 A0A1A9WQ28 W8AZD7 A0A1A9YIE8 A0A1A9UG79 B4J2Z7 A0A154NYU4 A0A1I8Q8J0 A0A0Q9WTA2 A0A1B0AQX4 A0A0L0CDE5 B4LFI5 B4PHG2 A0A195FUU5 A0A1W4VKE4 B3NHG2 Q9VU80 A0A0L7R825 A0A182UJF7 A0A182KST7 B4HH71 B4QJF9 A0A1S4HE00 A0A0P4VLC1 A0A232F012 E2B8S2 A0A0M4EHX2 B4MNC5 A0A151X9W6 B3MB37 F4WM09 B4KX60 A0A195BC47 A0A1W4VJ08 A0A023F303 A0A0A1XAU0 A0A0K8VAE1 A0A0R3P6F5 Q29E18 A0A0Q9XDW2 B4H423 A0A182JEY7 W5J591 Q8IQK0 A0A0A9YY25 A0A0Q9WWT4 A0A336LIR5 B0WEK1 A0A182H6A6 A0A1W4V6V8 A0A0M9ACA6 M9PI51 A0A0J9RUX6 A0A0Q5U824 A0A310S9S3 A0A182RGL3 A0A0Q9WK53 A0A0R1DZ98 A0A182HPY6 A0A182XDA3 A0A182FUF5 A0A0P8XWC3 A0A067R3G5 A0A182W9A0 A0A195C660 A0A0P8YJB2 A0A182PBM7 A0A182QDZ6 A0A146L973
A0A1W4W8F6 D6WHC9 A0A0C9QZX4 K7IXZ8 N6U264 A0A2P8XIA8 A0A3L8DY15 Q17M44 A0A158N9C2 A0A1B6E338 A0A2J7PCW7 A0A1B0C9F7 T1P9R3 A0A1B0FPK4 A0A1A9ZRW6 A0A0Q9XLX8 E0VL47 A0A0A1WGE2 A0A0K8VYC3 A0A026WC91 Q8IQK1 A0A1I8MZT9 A0A1A9WQ28 W8AZD7 A0A1A9YIE8 A0A1A9UG79 B4J2Z7 A0A154NYU4 A0A1I8Q8J0 A0A0Q9WTA2 A0A1B0AQX4 A0A0L0CDE5 B4LFI5 B4PHG2 A0A195FUU5 A0A1W4VKE4 B3NHG2 Q9VU80 A0A0L7R825 A0A182UJF7 A0A182KST7 B4HH71 B4QJF9 A0A1S4HE00 A0A0P4VLC1 A0A232F012 E2B8S2 A0A0M4EHX2 B4MNC5 A0A151X9W6 B3MB37 F4WM09 B4KX60 A0A195BC47 A0A1W4VJ08 A0A023F303 A0A0A1XAU0 A0A0K8VAE1 A0A0R3P6F5 Q29E18 A0A0Q9XDW2 B4H423 A0A182JEY7 W5J591 Q8IQK0 A0A0A9YY25 A0A0Q9WWT4 A0A336LIR5 B0WEK1 A0A182H6A6 A0A1W4V6V8 A0A0M9ACA6 M9PI51 A0A0J9RUX6 A0A0Q5U824 A0A310S9S3 A0A182RGL3 A0A0Q9WK53 A0A0R1DZ98 A0A182HPY6 A0A182XDA3 A0A182FUF5 A0A0P8XWC3 A0A067R3G5 A0A182W9A0 A0A195C660 A0A0P8YJB2 A0A182PBM7 A0A182QDZ6 A0A146L973
PDB
1WRM
E-value=6.05348e-49,
Score=487
Ontologies
GO
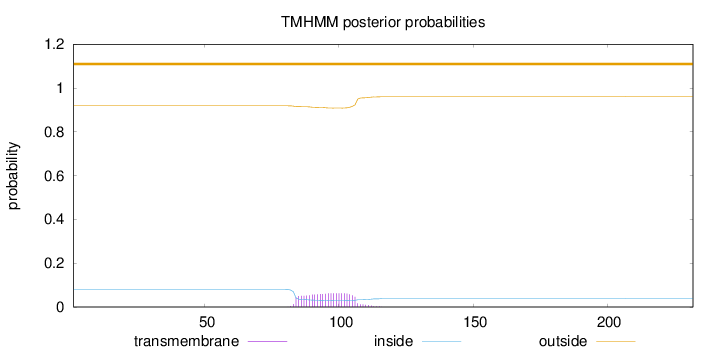
Topology
Length:
232
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.39942
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08093
outside
1 - 232
Population Genetic Test Statistics
Pi
219.048362
Theta
171.317463
Tajima's D
0.80221
CLR
0.291267
CSRT
0.597970101494925
Interpretation
Uncertain
