Gene
KWMTBOMO06550
Annotation
PREDICTED:_transmembrane_protein_184B_isoform_X1_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.708
Sequence
CDS
ATGAGTACCACAGATACGGTGAATATAACCGCTCAGCGGATTTCTGAAATTGTTCACGACGTGCTGAAGCCGCTGCAAGAGCCCATATTCCTGCAGACCCGCAGCGCGCAGGTCATCGCGGGAATATTCGTGTTCACGGCTCTATTCATTACTTGCCAACAGATATACCAGCATTTGCGTTGGTACACAAACCCATCAGAGCAACGTTGGATCGTGCGAATCCTATTTATCGTCCCAATATACGGCTGTTACAGCTGGATATCTTTGCTCTTCTTCAATGGAGACTCTTACTACGTTTACTTCTTCACCGTCAGGGATTGTTATGAAGCGTTCGTGATCTATAGCTTCTTGTCCCTGTGCTACGAGTACCTCGGGGGCGAAGGGAACATAATGTCGGAGTTGCGCGGGCGGCCGGTGCGCGCGTCCTGCGTCAACGGCACGTGTTGCCTGGCAGGAGCAACGTACACGATCGGCTTCCTCCGTTTCTGCAAGCAGGCAACCCTACAGTTCTGCCTCATAAAGCCGCTCTGCGCTTTCGTCATCATTTTCTTGCAGGCTTTCGGGCATTACCACGACGGTGACTGGAGTCCCAACGGCGGCTACATTTACATAACGATCATCAATAATTTCTCCGTGAGCTTGGCACTCTACGGCCTGTTCCTCTTCCTCGGAGCCACCAGGGAGATGCTGAAGCCTTTCGATCCAGTTCTGAAGTTCTTCACCGTTAAATCTGTCATCTTCCTTTCGTTCTGGCAAGGCGTCGCCCTGGCCATACTCGAGAAGGCGCAAGTGATCTCCCCGATACTGGACGCGAGCAACGTTCCCACGACGGCCGGTACTGTGTCGGCCGGCTACCAGAACTTCCTGATCTGCATCGAGATGTGCGCCGCGGCGGTGGCGCTGCGGTACGCGTTCCCGGCCGCCGTGTACGTGCACTCGCACCGCGACCCGCACCGCTCCCTCACCATGCAGTCGATCTCCAGCAGCCTCAAGGAGACAATGAATCCCAAGGACATAATGACGGACGCCTTCCACAATTTCCATCCGCAGTACCAGCAGTACACCCAGTACAGCTCCGTAATGCGCGGTCACACGGAATCCGGTAAGCGTGGGGAGGAAGGAGCGGCTCTGGCTCCTCGCCCGCCGCCCCAGCGCGTCGCTACCATCTGCCACGCCACCAATTACCACGAGAAGACTACGCTGCTCAGCTCCGATGACGAATTTCAGTGA
Protein
MSTTDTVNITAQRISEIVHDVLKPLQEPIFLQTRSAQVIAGIFVFTALFITCQQIYQHLRWYTNPSEQRWIVRILFIVPIYGCYSWISLLFFNGDSYYVYFFTVRDCYEAFVIYSFLSLCYEYLGGEGNIMSELRGRPVRASCVNGTCCLAGATYTIGFLRFCKQATLQFCLIKPLCAFVIIFLQAFGHYHDGDWSPNGGYIYITIINNFSVSLALYGLFLFLGATREMLKPFDPVLKFFTVKSVIFLSFWQGVALAILEKAQVISPILDASNVPTTAGTVSAGYQNFLICIEMCAAAVALRYAFPAAVYVHSHRDPHRSLTMQSISSSLKETMNPKDIMTDAFHNFHPQYQQYTQYSSVMRGHTESGKRGEEGAALAPRPPPQRVATICHATNYHEKTTLLSSDDEFQ
Summary
Uniprot
A0A212EQT9
A0A2W1BTU5
A0A2H1V9Q4
A0A2A4J0V3
A0A194PW99
A0A194QQL9
+ More
S4P291 A0A026W211 E9IGT2 E2BBR8 E2AVY5 A0A232F6J8 A0A158NDX8 A0A195ESB7 A0A151X4H3 A0A195BR99 A0A195C4Y1 K7J338 A0A195D7Y3 A0A087ZW91 V9I992 A0A1L8DJV6 E0VYV2 A0A0Q9WII3 A0A310SBD2 A0A1I8PB65 A0A0L7QV55 A0A0Q9XD69 A0A1B6C1K0 A0A1I8MCW3 A0A3B0JS68 A0A0M4EA54 A0A1A9WQ14 A0A0R3P479 W8B2N9 U4UN65 A0A0A1WKT1 A0A0K8UAP1 A0A1W4W7T6 A0A1Y1KZ84 A0A0P9AKR9 A0A0J9RLU2 A1A714 A0A1Q3EUK1 A0A067RKV2 B4N2V1 B4IZJ8 B4LHM2 A0A1Q3EUA7 Q17AK3 T1E1I4 A0A023ESF5 A0A1B6JZF4 A0A1I8PB57 A0A1L8EBH3 B4KWA2 A0A084VDN3 A0A336KYG5 A0A1L8DJV2 A0A1L8DK90 A0A2M4BQ87 A0A2M4ALB4 W5JIR3 A0A2M3Z5L2 Q7PNZ7 A0A1I8MCW6 A0A182IZ47 A0A3B0JS73 A0A1A9V4F5 A0A1A9ZLX2 A0A1A9YG16 W8C149 A0A1B6D6W4 A0A0Q5UGX1 D6WF17 A0A1W4WL99 A0A034W2W9 A0A0K8WID9 A0A1Y1KXV4 U5EWE1 V9I7V7 Q2LZX6 B4HBR3 A0A0K8VWX7 A0A0R1DUB7 A0A0A1X2A9 B3MAZ5 B3NBC8 B4PDA8 B4HVV4 B4QMG6 Q960F6 A0A1W4VVC9 A0A1J1I7J5 A0A1B0CP60 A0A0K8TNW0 A0A2S2QNM9 A0A1B6MFA8 D3TQ11 A0A0A9XHN3
S4P291 A0A026W211 E9IGT2 E2BBR8 E2AVY5 A0A232F6J8 A0A158NDX8 A0A195ESB7 A0A151X4H3 A0A195BR99 A0A195C4Y1 K7J338 A0A195D7Y3 A0A087ZW91 V9I992 A0A1L8DJV6 E0VYV2 A0A0Q9WII3 A0A310SBD2 A0A1I8PB65 A0A0L7QV55 A0A0Q9XD69 A0A1B6C1K0 A0A1I8MCW3 A0A3B0JS68 A0A0M4EA54 A0A1A9WQ14 A0A0R3P479 W8B2N9 U4UN65 A0A0A1WKT1 A0A0K8UAP1 A0A1W4W7T6 A0A1Y1KZ84 A0A0P9AKR9 A0A0J9RLU2 A1A714 A0A1Q3EUK1 A0A067RKV2 B4N2V1 B4IZJ8 B4LHM2 A0A1Q3EUA7 Q17AK3 T1E1I4 A0A023ESF5 A0A1B6JZF4 A0A1I8PB57 A0A1L8EBH3 B4KWA2 A0A084VDN3 A0A336KYG5 A0A1L8DJV2 A0A1L8DK90 A0A2M4BQ87 A0A2M4ALB4 W5JIR3 A0A2M3Z5L2 Q7PNZ7 A0A1I8MCW6 A0A182IZ47 A0A3B0JS73 A0A1A9V4F5 A0A1A9ZLX2 A0A1A9YG16 W8C149 A0A1B6D6W4 A0A0Q5UGX1 D6WF17 A0A1W4WL99 A0A034W2W9 A0A0K8WID9 A0A1Y1KXV4 U5EWE1 V9I7V7 Q2LZX6 B4HBR3 A0A0K8VWX7 A0A0R1DUB7 A0A0A1X2A9 B3MAZ5 B3NBC8 B4PDA8 B4HVV4 B4QMG6 Q960F6 A0A1W4VVC9 A0A1J1I7J5 A0A1B0CP60 A0A0K8TNW0 A0A2S2QNM9 A0A1B6MFA8 D3TQ11 A0A0A9XHN3
Pubmed
22118469
28756777
26354079
23622113
24508170
30249741
+ More
21282665 20798317 28648823 21347285 20075255 20566863 17994087 18057021 25315136 15632085 24495485 23537049 25830018 28004739 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 17510324 24330624 24945155 26483478 24438588 20920257 23761445 12364791 14747013 17210077 18362917 19820115 25348373 17550304 26369729 20353571 25401762
21282665 20798317 28648823 21347285 20075255 20566863 17994087 18057021 25315136 15632085 24495485 23537049 25830018 28004739 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 17510324 24330624 24945155 26483478 24438588 20920257 23761445 12364791 14747013 17210077 18362917 19820115 25348373 17550304 26369729 20353571 25401762
EMBL
AGBW02013252
OWR43821.1
KZ149971
PZC76056.1
ODYU01001414
SOQ37559.1
+ More
NWSH01004467 PCG65070.1 KQ459590 KPI97278.1 KQ461190 KPJ07265.1 GAIX01008596 JAA83964.1 KK107503 QOIP01000006 EZA49611.1 RLU21344.1 GL763054 EFZ20245.1 GL447151 EFN86844.1 GL443213 EFN62407.1 NNAY01000884 OXU26068.1 ADTU01013062 KQ981993 KYN30797.1 KQ982548 KYQ55287.1 KQ976424 KYM88486.1 KQ978251 KYM95929.1 KQ981153 KYN08981.1 JR036530 JR036533 JR036534 JR036536 AEY57201.1 AEY57204.1 AEY57205.1 AEY57207.1 GFDF01007358 JAV06726.1 DS235848 EEB18558.1 CH940647 KRF84457.1 KQ776210 OAD52204.1 KQ414727 KOC62523.1 CH933809 KRG06229.1 KRG06230.1 GEDC01029930 JAS07368.1 OUUW01000009 SPP84974.1 CP012525 ALC44091.1 CH379069 KRT07929.1 GAMC01011145 JAB95410.1 KB632292 ERL91606.1 GBXI01014830 JAC99461.1 GDHF01028610 JAI23704.1 GEZM01075223 JAV64197.1 CH902618 KPU78465.1 KPU78466.1 CM002912 KMY96871.1 KMY96872.1 BT029687 AE014296 ABL75744.1 ABW08437.1 GFDL01016046 JAV18999.1 KK852567 KDR21220.1 CH964062 EDW78690.2 CH916366 EDV97773.1 EDW69575.1 GFDL01016146 JAV18899.1 CH477333 EAT43288.1 GALA01001634 JAA93218.1 JXUM01034619 JXUM01034620 JXUM01034621 JXUM01034622 JXUM01034623 JXUM01034624 JXUM01034625 JXUM01034626 JXUM01034627 JXUM01034628 GAPW01001907 JAC11691.1 GECU01003131 JAT04576.1 GFDG01002896 JAV15903.1 EDW18509.1 ATLV01011631 KE524705 KFB36077.1 UFQS01001036 UFQT01001036 SSX08663.1 SSX28578.1 GFDF01007325 JAV06759.1 GFDF01007324 JAV06760.1 GGFJ01006041 MBW55182.1 GGFK01008201 MBW41522.1 ADMH02001054 ETN64272.1 GGFM01003060 MBW23811.1 AAAB01008960 EAA11279.5 SPP84975.1 GAMC01011146 JAB95409.1 GEDC01022120 GEDC01015885 GEDC01006787 JAS15178.1 JAS21413.1 JAS30511.1 CH954178 KQS43088.1 KQ971319 EFA00467.2 GAKP01010834 JAC48118.1 GDHF01001674 JAI50640.1 GEZM01075224 JAV64196.1 GANO01002987 JAB56884.1 JR036529 JR036531 JR036532 JR036535 AEY57200.1 AEY57202.1 AEY57203.1 AEY57206.1 EAL31161.1 CH479261 EDW39467.1 GDHF01013898 GDHF01008931 JAI38416.1 JAI43383.1 CM000159 KRK00763.1 GBXI01008813 JAD05479.1 EDV40261.1 KPU78467.1 EDV50181.1 EDW92856.1 CH480817 EDW50069.1 CM000363 EDX08823.1 KMY96870.1 AY052085 AAF47516.2 AAK93509.1 CVRI01000043 CRK96232.1 AJWK01021443 AJWK01021444 AJWK01021445 AJWK01021446 GDAI01001549 JAI16054.1 GGMS01010118 MBY79321.1 GEBQ01005402 JAT34575.1 CCAG010008841 EZ423513 ADD19789.1 GBHO01024473 JAG19131.1
NWSH01004467 PCG65070.1 KQ459590 KPI97278.1 KQ461190 KPJ07265.1 GAIX01008596 JAA83964.1 KK107503 QOIP01000006 EZA49611.1 RLU21344.1 GL763054 EFZ20245.1 GL447151 EFN86844.1 GL443213 EFN62407.1 NNAY01000884 OXU26068.1 ADTU01013062 KQ981993 KYN30797.1 KQ982548 KYQ55287.1 KQ976424 KYM88486.1 KQ978251 KYM95929.1 KQ981153 KYN08981.1 JR036530 JR036533 JR036534 JR036536 AEY57201.1 AEY57204.1 AEY57205.1 AEY57207.1 GFDF01007358 JAV06726.1 DS235848 EEB18558.1 CH940647 KRF84457.1 KQ776210 OAD52204.1 KQ414727 KOC62523.1 CH933809 KRG06229.1 KRG06230.1 GEDC01029930 JAS07368.1 OUUW01000009 SPP84974.1 CP012525 ALC44091.1 CH379069 KRT07929.1 GAMC01011145 JAB95410.1 KB632292 ERL91606.1 GBXI01014830 JAC99461.1 GDHF01028610 JAI23704.1 GEZM01075223 JAV64197.1 CH902618 KPU78465.1 KPU78466.1 CM002912 KMY96871.1 KMY96872.1 BT029687 AE014296 ABL75744.1 ABW08437.1 GFDL01016046 JAV18999.1 KK852567 KDR21220.1 CH964062 EDW78690.2 CH916366 EDV97773.1 EDW69575.1 GFDL01016146 JAV18899.1 CH477333 EAT43288.1 GALA01001634 JAA93218.1 JXUM01034619 JXUM01034620 JXUM01034621 JXUM01034622 JXUM01034623 JXUM01034624 JXUM01034625 JXUM01034626 JXUM01034627 JXUM01034628 GAPW01001907 JAC11691.1 GECU01003131 JAT04576.1 GFDG01002896 JAV15903.1 EDW18509.1 ATLV01011631 KE524705 KFB36077.1 UFQS01001036 UFQT01001036 SSX08663.1 SSX28578.1 GFDF01007325 JAV06759.1 GFDF01007324 JAV06760.1 GGFJ01006041 MBW55182.1 GGFK01008201 MBW41522.1 ADMH02001054 ETN64272.1 GGFM01003060 MBW23811.1 AAAB01008960 EAA11279.5 SPP84975.1 GAMC01011146 JAB95409.1 GEDC01022120 GEDC01015885 GEDC01006787 JAS15178.1 JAS21413.1 JAS30511.1 CH954178 KQS43088.1 KQ971319 EFA00467.2 GAKP01010834 JAC48118.1 GDHF01001674 JAI50640.1 GEZM01075224 JAV64196.1 GANO01002987 JAB56884.1 JR036529 JR036531 JR036532 JR036535 AEY57200.1 AEY57202.1 AEY57203.1 AEY57206.1 EAL31161.1 CH479261 EDW39467.1 GDHF01013898 GDHF01008931 JAI38416.1 JAI43383.1 CM000159 KRK00763.1 GBXI01008813 JAD05479.1 EDV40261.1 KPU78467.1 EDV50181.1 EDW92856.1 CH480817 EDW50069.1 CM000363 EDX08823.1 KMY96870.1 AY052085 AAF47516.2 AAK93509.1 CVRI01000043 CRK96232.1 AJWK01021443 AJWK01021444 AJWK01021445 AJWK01021446 GDAI01001549 JAI16054.1 GGMS01010118 MBY79321.1 GEBQ01005402 JAT34575.1 CCAG010008841 EZ423513 ADD19789.1 GBHO01024473 JAG19131.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000053097
UP000279307
+ More
UP000008237 UP000000311 UP000215335 UP000005205 UP000078541 UP000075809 UP000078540 UP000078542 UP000002358 UP000078492 UP000005203 UP000009046 UP000008792 UP000095300 UP000053825 UP000009192 UP000095301 UP000268350 UP000092553 UP000091820 UP000001819 UP000030742 UP000192221 UP000007801 UP000000803 UP000027135 UP000007798 UP000001070 UP000008820 UP000069940 UP000030765 UP000000673 UP000007062 UP000075880 UP000078200 UP000092445 UP000092443 UP000008711 UP000007266 UP000192223 UP000008744 UP000002282 UP000001292 UP000000304 UP000183832 UP000092461 UP000092444
UP000008237 UP000000311 UP000215335 UP000005205 UP000078541 UP000075809 UP000078540 UP000078542 UP000002358 UP000078492 UP000005203 UP000009046 UP000008792 UP000095300 UP000053825 UP000009192 UP000095301 UP000268350 UP000092553 UP000091820 UP000001819 UP000030742 UP000192221 UP000007801 UP000000803 UP000027135 UP000007798 UP000001070 UP000008820 UP000069940 UP000030765 UP000000673 UP000007062 UP000075880 UP000078200 UP000092445 UP000092443 UP000008711 UP000007266 UP000192223 UP000008744 UP000002282 UP000001292 UP000000304 UP000183832 UP000092461 UP000092444
SUPFAM
SSF63411
SSF63411
ProteinModelPortal
A0A212EQT9
A0A2W1BTU5
A0A2H1V9Q4
A0A2A4J0V3
A0A194PW99
A0A194QQL9
+ More
S4P291 A0A026W211 E9IGT2 E2BBR8 E2AVY5 A0A232F6J8 A0A158NDX8 A0A195ESB7 A0A151X4H3 A0A195BR99 A0A195C4Y1 K7J338 A0A195D7Y3 A0A087ZW91 V9I992 A0A1L8DJV6 E0VYV2 A0A0Q9WII3 A0A310SBD2 A0A1I8PB65 A0A0L7QV55 A0A0Q9XD69 A0A1B6C1K0 A0A1I8MCW3 A0A3B0JS68 A0A0M4EA54 A0A1A9WQ14 A0A0R3P479 W8B2N9 U4UN65 A0A0A1WKT1 A0A0K8UAP1 A0A1W4W7T6 A0A1Y1KZ84 A0A0P9AKR9 A0A0J9RLU2 A1A714 A0A1Q3EUK1 A0A067RKV2 B4N2V1 B4IZJ8 B4LHM2 A0A1Q3EUA7 Q17AK3 T1E1I4 A0A023ESF5 A0A1B6JZF4 A0A1I8PB57 A0A1L8EBH3 B4KWA2 A0A084VDN3 A0A336KYG5 A0A1L8DJV2 A0A1L8DK90 A0A2M4BQ87 A0A2M4ALB4 W5JIR3 A0A2M3Z5L2 Q7PNZ7 A0A1I8MCW6 A0A182IZ47 A0A3B0JS73 A0A1A9V4F5 A0A1A9ZLX2 A0A1A9YG16 W8C149 A0A1B6D6W4 A0A0Q5UGX1 D6WF17 A0A1W4WL99 A0A034W2W9 A0A0K8WID9 A0A1Y1KXV4 U5EWE1 V9I7V7 Q2LZX6 B4HBR3 A0A0K8VWX7 A0A0R1DUB7 A0A0A1X2A9 B3MAZ5 B3NBC8 B4PDA8 B4HVV4 B4QMG6 Q960F6 A0A1W4VVC9 A0A1J1I7J5 A0A1B0CP60 A0A0K8TNW0 A0A2S2QNM9 A0A1B6MFA8 D3TQ11 A0A0A9XHN3
S4P291 A0A026W211 E9IGT2 E2BBR8 E2AVY5 A0A232F6J8 A0A158NDX8 A0A195ESB7 A0A151X4H3 A0A195BR99 A0A195C4Y1 K7J338 A0A195D7Y3 A0A087ZW91 V9I992 A0A1L8DJV6 E0VYV2 A0A0Q9WII3 A0A310SBD2 A0A1I8PB65 A0A0L7QV55 A0A0Q9XD69 A0A1B6C1K0 A0A1I8MCW3 A0A3B0JS68 A0A0M4EA54 A0A1A9WQ14 A0A0R3P479 W8B2N9 U4UN65 A0A0A1WKT1 A0A0K8UAP1 A0A1W4W7T6 A0A1Y1KZ84 A0A0P9AKR9 A0A0J9RLU2 A1A714 A0A1Q3EUK1 A0A067RKV2 B4N2V1 B4IZJ8 B4LHM2 A0A1Q3EUA7 Q17AK3 T1E1I4 A0A023ESF5 A0A1B6JZF4 A0A1I8PB57 A0A1L8EBH3 B4KWA2 A0A084VDN3 A0A336KYG5 A0A1L8DJV2 A0A1L8DK90 A0A2M4BQ87 A0A2M4ALB4 W5JIR3 A0A2M3Z5L2 Q7PNZ7 A0A1I8MCW6 A0A182IZ47 A0A3B0JS73 A0A1A9V4F5 A0A1A9ZLX2 A0A1A9YG16 W8C149 A0A1B6D6W4 A0A0Q5UGX1 D6WF17 A0A1W4WL99 A0A034W2W9 A0A0K8WID9 A0A1Y1KXV4 U5EWE1 V9I7V7 Q2LZX6 B4HBR3 A0A0K8VWX7 A0A0R1DUB7 A0A0A1X2A9 B3MAZ5 B3NBC8 B4PDA8 B4HVV4 B4QMG6 Q960F6 A0A1W4VVC9 A0A1J1I7J5 A0A1B0CP60 A0A0K8TNW0 A0A2S2QNM9 A0A1B6MFA8 D3TQ11 A0A0A9XHN3
Ontologies
PANTHER
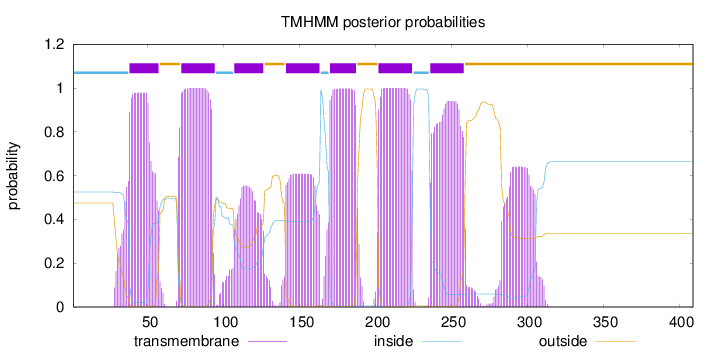
Topology
Length:
409
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
146.74581
Exp number, first 60 AAs:
20.34108
Total prob of N-in:
0.52492
POSSIBLE N-term signal
sequence
inside
1 - 37
TMhelix
38 - 57
outside
58 - 71
TMhelix
72 - 94
inside
95 - 106
TMhelix
107 - 126
outside
127 - 140
TMhelix
141 - 163
inside
164 - 169
TMhelix
170 - 187
outside
188 - 201
TMhelix
202 - 224
inside
225 - 235
TMhelix
236 - 258
outside
259 - 409
Population Genetic Test Statistics
Pi
22.863864
Theta
170.210315
Tajima's D
0.720272
CLR
0.408296
CSRT
0.576021198940053
Interpretation
Uncertain
