Gene
KWMTBOMO06546
Pre Gene Modal
BGIBMGA011649
Annotation
PREDICTED:_uncharacterized_protein_LOC106134869_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.57
Sequence
CDS
ATGCCGTCAGGGGTTATATTCGTTGTTTGTATACCCTCAAGTAATTATGAATATATTTTAAAATTCGGAAAACCTAAGGATTTAATAGACATTAATGATTTCGCACCAAAACCCTTTCGCGTTGCTTTGGAATTGAGGGGCAAGAGATCCTTAAACATTCCTGTACATTTGACACAAGAACAATATTGCGATCAAGCATTCACTCAATTGGAGAGTATTAAAGAAGAACATTTTTTTACAATGGAGAACATTTTACAAGAACTTTTGAAAAAACTTCGCATAGAACACGTAGTGTGGGTGAGTGATAAAGTGGGCAATTATCACGAGGTCATATTCCCTGTTAAAACCGGGGACCAATGCGAAACGTTACTTCATTGTTTAACGCAACTTGGTATCGGTGTCAAATGTAAATCTATAGTGAGTGTTTTGCCATGTAGTGTCGTGCACTCGGCAATCGATGATGATCTAAATGACGATGAAACCATATTGCGCCAAACAGAAGACGCGAAAAGATGGAGAAGTTTCGTTGAATCAATAAGGTCAAAATTGACAGTGAAACAAGTAGTAGACGGAGTTCGCGGAGGTGGAGAGTTATCATTTGATTATCTTACTCTTATCTTAACAGCTGACTCTCTGGCAGCTTTGGGTCTTGTTGAAAATAACGCGTCTAATATAGTAGCGGCAATGCTGGTATCTCCTTTAATGGGTCCGGTGATGTCTATAACTTTCGGCACAATAATAGCCGATAGGAGTCTAATAAGGAGCGGTTTTGAAAGTCTAATTGTGGGCATGATCTTCTCTCTGATTTTTGGTTTCATATTTGGATTGATATTGGGAACGACTGAAATGCCTTGGGGATTTGGTGATTGGCCAACAGAAGAAATGAAATCACGAGGCAACGTACGATCACTTTGGATGGGCATTTTATGGGCACTAACTTCTGGAACTGGTGTAGCATTAGCTCTACTACAAGGCAGCGCGGGTCCATTAATCGGTATAGCTATTTCAGCATCTTTATTGCCACCAGTGGTGAATTGTGGTTTATTTTGGGGGTTGGCATGTATTTGGCTGCTATATCCAGGGATAAAAATGCCTCATATAAAAGGAGAATCTTACTTTGGAAACTCGTCATATCAACCTTTGTATCACGATTATATTCCAATAGAATTCGCCGTTAATGGTATTGTTAGTTGTTGTCTAACCATCGTAAACGTTATTTGTATTTTTATAACCGCCATAATATTTCTGAAGATAAAAGAAGTAGCAGCGCCCTATACTTCCAGCCCGGATTTAAGAAGATTTTGGGAGCAAGATATCAAAGTTGCTAGAGAAGCTAACAAATCTAATTTGCGAGACGAAGATGACGAAAGAACCGAATTAGTTTTAGACGACCTTAAAGAAACAGACGAAAGCATGAAGGAAAAATTAGAAGCAGCCGTCGAAGAGGCTCTGAATGACGAAACGTACAGGAAAGTGAAAAGAATGAGTTATCAAAGTCACAACGCCGACGAGATTGGCAGAACTATTGGACACCAAACTTCAAAACGATCTAGTATTTATTCAGATGCCTCTACGCCGGTAACAAAAGCTAACAGTGAAGACATAGCCGCATTGGATAAACTGCTTACGTCGATTCTCGGGATCAAGGATATCAACCGAAGATCATTTATTTCGCAACGTGATGCTTCGAGGATCAATAACATGCCCTCTATTGCGGAATTGGGGAACAGACGATCAGAATCTACACCCGATCGCATTGCGGAAGACTCTATAATAATGAGCGTAATAAATAATTTGAAAACAAATATGAATAACAGTTCTTCTGACGAAACCTTTCTAACACCTGCATGA
Protein
MPSGVIFVVCIPSSNYEYILKFGKPKDLIDINDFAPKPFRVALELRGKRSLNIPVHLTQEQYCDQAFTQLESIKEEHFFTMENILQELLKKLRIEHVVWVSDKVGNYHEVIFPVKTGDQCETLLHCLTQLGIGVKCKSIVSVLPCSVVHSAIDDDLNDDETILRQTEDAKRWRSFVESIRSKLTVKQVVDGVRGGGELSFDYLTLILTADSLAALGLVENNASNIVAAMLVSPLMGPVMSITFGTIIADRSLIRSGFESLIVGMIFSLIFGFIFGLILGTTEMPWGFGDWPTEEMKSRGNVRSLWMGILWALTSGTGVALALLQGSAGPLIGIAISASLLPPVVNCGLFWGLACIWLLYPGIKMPHIKGESYFGNSSYQPLYHDYIPIEFAVNGIVSCCLTIVNVICIFITAIIFLKIKEVAAPYTSSPDLRRFWEQDIKVAREANKSNLRDEDDERTELVLDDLKETDESMKEKLEAAVEEALNDETYRKVKRMSYQSHNADEIGRTIGHQTSKRSSIYSDASTPVTKANSEDIAALDKLLTSILGIKDINRRSFISQRDASRINNMPSIAELGNRRSESTPDRIAEDSIIMSVINNLKTNMNNSSSDETFLTPA
Summary
Uniprot
A0A2A4JNZ3
A0A2H1VBF9
A0A212EQP2
A0A194PWY5
A0A194QQM5
A0A023EUX2
+ More
A0A084WG70 A0A2C9H3W9 A0A2C9GPZ1 A0A336M6G2 A0A1A9UPM2 A0A1A9WKE5 E9I9V5 A0A1B0AT89 A0A182VA31 A0A1A9XN50 A0A182YDF4 A0A2A3EPB9 A0A0L7QT62 A0A151X1I7 A0A087ZVR1 W5JBZ8 F4W457 A0A195B3E3 A0A158NF08 A0A026WLQ4 A0A195E4Q9 A0A182REY6 A0A182WGM9 A0A195ETK1 A0A0M9A7J5 A0A1I8Q9C9 A0A195CJQ1 T1PBL0 A0A1I8MZI5 A0A3L8D5X1 A0A3B0KBR4 Q0IGY6 B4NLH9 A0A0J9TK34 A0A0R1DSG0 B3MPL2 B3N924 A0A1W4UF67 B4HWG0 E2B6P8 A0A0Q9XG44 B4M9D1 Q29NH1 E2AT06 A0A182PCT9 A0A182NBV4 A0A0K8U4R8 W8B581 A0A1B0AJS0 A0A0A1X8K9 A0A310SKX7 A0A034VW02 A0A0K8VIQ7 E0VVU9 A0A2P8XZJ7 A0A293MY73 A0A087SXH2 A0A1Z5L5A3 A0A1Z5KVR4 A0A1W4XUS7 U5EUZ1
A0A084WG70 A0A2C9H3W9 A0A2C9GPZ1 A0A336M6G2 A0A1A9UPM2 A0A1A9WKE5 E9I9V5 A0A1B0AT89 A0A182VA31 A0A1A9XN50 A0A182YDF4 A0A2A3EPB9 A0A0L7QT62 A0A151X1I7 A0A087ZVR1 W5JBZ8 F4W457 A0A195B3E3 A0A158NF08 A0A026WLQ4 A0A195E4Q9 A0A182REY6 A0A182WGM9 A0A195ETK1 A0A0M9A7J5 A0A1I8Q9C9 A0A195CJQ1 T1PBL0 A0A1I8MZI5 A0A3L8D5X1 A0A3B0KBR4 Q0IGY6 B4NLH9 A0A0J9TK34 A0A0R1DSG0 B3MPL2 B3N924 A0A1W4UF67 B4HWG0 E2B6P8 A0A0Q9XG44 B4M9D1 Q29NH1 E2AT06 A0A182PCT9 A0A182NBV4 A0A0K8U4R8 W8B581 A0A1B0AJS0 A0A0A1X8K9 A0A310SKX7 A0A034VW02 A0A0K8VIQ7 E0VVU9 A0A2P8XZJ7 A0A293MY73 A0A087SXH2 A0A1Z5L5A3 A0A1Z5KVR4 A0A1W4XUS7 U5EUZ1
Pubmed
22118469
26354079
24945155
24438588
12364791
21282665
+ More
25244985 20920257 23761445 21719571 21347285 24508170 25315136 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 17550304 18057021 20798317 15632085 24495485 25830018 25348373 20566863 29403074 28528879
25244985 20920257 23761445 21719571 21347285 24508170 25315136 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 22936249 17550304 18057021 20798317 15632085 24495485 25830018 25348373 20566863 29403074 28528879
EMBL
NWSH01001010
PCG73140.1
ODYU01001414
SOQ37564.1
AGBW02013252
OWR43816.1
+ More
KQ459590 KPI97274.1 KQ461190 KPJ07270.1 GAPW01000358 JAC13240.1 ATLV01023453 ATLV01023454 ATLV01023455 ATLV01023456 KE525343 KFB49214.1 AAAB01008980 APCN01001744 UFQT01000449 SSX24469.1 GL761846 EFZ22636.1 JXJN01003204 JXJN01003205 KZ288210 PBC32979.1 KQ414755 KOC61744.1 KQ982585 KYQ54283.1 ADMH02001647 ETN61561.1 GL887491 EGI71053.1 KQ976625 KYM79013.1 ADTU01001499 KK107153 EZA56982.1 KQ979609 KYN20148.1 KQ981979 KYN31486.1 KQ435727 KOX78076.1 KQ977642 KYN00956.1 KA645495 AFP60124.1 QOIP01000012 RLU15810.1 OUUW01000010 SPP85610.1 AE014134 BT028784 ABI31305.2 ABI34165.1 CH964274 EDW85218.2 CM002910 KMY89400.1 CM000157 KRJ97993.1 CH902620 EDV32260.2 KPU73863.1 CH954177 EDV58459.2 CH480818 EDW52355.1 GL446000 EFN88632.1 CH933807 KRG03169.1 CH940654 EDW57807.2 KRF77964.1 CH379060 EAL33371.3 GL442439 EFN63441.1 GDHF01031019 JAI21295.1 GAMC01012803 GAMC01012802 JAB93752.1 GBXI01007217 JAD07075.1 KQ760208 OAD61381.1 GAKP01012685 JAC46267.1 GDHF01013864 JAI38450.1 DS235815 EEB17505.1 PYGN01001120 PSN37428.1 GFWV01020728 MAA45456.1 KK112401 KFM57561.1 GFJQ02004495 JAW02475.1 GFJQ02007791 JAV99178.1 GANO01003649 JAB56222.1
KQ459590 KPI97274.1 KQ461190 KPJ07270.1 GAPW01000358 JAC13240.1 ATLV01023453 ATLV01023454 ATLV01023455 ATLV01023456 KE525343 KFB49214.1 AAAB01008980 APCN01001744 UFQT01000449 SSX24469.1 GL761846 EFZ22636.1 JXJN01003204 JXJN01003205 KZ288210 PBC32979.1 KQ414755 KOC61744.1 KQ982585 KYQ54283.1 ADMH02001647 ETN61561.1 GL887491 EGI71053.1 KQ976625 KYM79013.1 ADTU01001499 KK107153 EZA56982.1 KQ979609 KYN20148.1 KQ981979 KYN31486.1 KQ435727 KOX78076.1 KQ977642 KYN00956.1 KA645495 AFP60124.1 QOIP01000012 RLU15810.1 OUUW01000010 SPP85610.1 AE014134 BT028784 ABI31305.2 ABI34165.1 CH964274 EDW85218.2 CM002910 KMY89400.1 CM000157 KRJ97993.1 CH902620 EDV32260.2 KPU73863.1 CH954177 EDV58459.2 CH480818 EDW52355.1 GL446000 EFN88632.1 CH933807 KRG03169.1 CH940654 EDW57807.2 KRF77964.1 CH379060 EAL33371.3 GL442439 EFN63441.1 GDHF01031019 JAI21295.1 GAMC01012803 GAMC01012802 JAB93752.1 GBXI01007217 JAD07075.1 KQ760208 OAD61381.1 GAKP01012685 JAC46267.1 GDHF01013864 JAI38450.1 DS235815 EEB17505.1 PYGN01001120 PSN37428.1 GFWV01020728 MAA45456.1 KK112401 KFM57561.1 GFJQ02004495 JAW02475.1 GFJQ02007791 JAV99178.1 GANO01003649 JAB56222.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000030765
UP000075840
+ More
UP000078200 UP000091820 UP000092460 UP000075903 UP000092443 UP000076408 UP000242457 UP000053825 UP000075809 UP000005203 UP000000673 UP000007755 UP000078540 UP000005205 UP000053097 UP000078492 UP000075900 UP000075920 UP000078541 UP000053105 UP000095300 UP000078542 UP000095301 UP000279307 UP000268350 UP000000803 UP000007798 UP000002282 UP000007801 UP000008711 UP000192221 UP000001292 UP000008237 UP000009192 UP000008792 UP000001819 UP000000311 UP000075885 UP000075884 UP000092445 UP000009046 UP000245037 UP000054359 UP000192223
UP000078200 UP000091820 UP000092460 UP000075903 UP000092443 UP000076408 UP000242457 UP000053825 UP000075809 UP000005203 UP000000673 UP000007755 UP000078540 UP000005205 UP000053097 UP000078492 UP000075900 UP000075920 UP000078541 UP000053105 UP000095300 UP000078542 UP000095301 UP000279307 UP000268350 UP000000803 UP000007798 UP000002282 UP000007801 UP000008711 UP000192221 UP000001292 UP000008237 UP000009192 UP000008792 UP000001819 UP000000311 UP000075885 UP000075884 UP000092445 UP000009046 UP000245037 UP000054359 UP000192223
PRIDE
Pfam
PF04087 DUF389
CDD
ProteinModelPortal
A0A2A4JNZ3
A0A2H1VBF9
A0A212EQP2
A0A194PWY5
A0A194QQM5
A0A023EUX2
+ More
A0A084WG70 A0A2C9H3W9 A0A2C9GPZ1 A0A336M6G2 A0A1A9UPM2 A0A1A9WKE5 E9I9V5 A0A1B0AT89 A0A182VA31 A0A1A9XN50 A0A182YDF4 A0A2A3EPB9 A0A0L7QT62 A0A151X1I7 A0A087ZVR1 W5JBZ8 F4W457 A0A195B3E3 A0A158NF08 A0A026WLQ4 A0A195E4Q9 A0A182REY6 A0A182WGM9 A0A195ETK1 A0A0M9A7J5 A0A1I8Q9C9 A0A195CJQ1 T1PBL0 A0A1I8MZI5 A0A3L8D5X1 A0A3B0KBR4 Q0IGY6 B4NLH9 A0A0J9TK34 A0A0R1DSG0 B3MPL2 B3N924 A0A1W4UF67 B4HWG0 E2B6P8 A0A0Q9XG44 B4M9D1 Q29NH1 E2AT06 A0A182PCT9 A0A182NBV4 A0A0K8U4R8 W8B581 A0A1B0AJS0 A0A0A1X8K9 A0A310SKX7 A0A034VW02 A0A0K8VIQ7 E0VVU9 A0A2P8XZJ7 A0A293MY73 A0A087SXH2 A0A1Z5L5A3 A0A1Z5KVR4 A0A1W4XUS7 U5EUZ1
A0A084WG70 A0A2C9H3W9 A0A2C9GPZ1 A0A336M6G2 A0A1A9UPM2 A0A1A9WKE5 E9I9V5 A0A1B0AT89 A0A182VA31 A0A1A9XN50 A0A182YDF4 A0A2A3EPB9 A0A0L7QT62 A0A151X1I7 A0A087ZVR1 W5JBZ8 F4W457 A0A195B3E3 A0A158NF08 A0A026WLQ4 A0A195E4Q9 A0A182REY6 A0A182WGM9 A0A195ETK1 A0A0M9A7J5 A0A1I8Q9C9 A0A195CJQ1 T1PBL0 A0A1I8MZI5 A0A3L8D5X1 A0A3B0KBR4 Q0IGY6 B4NLH9 A0A0J9TK34 A0A0R1DSG0 B3MPL2 B3N924 A0A1W4UF67 B4HWG0 E2B6P8 A0A0Q9XG44 B4M9D1 Q29NH1 E2AT06 A0A182PCT9 A0A182NBV4 A0A0K8U4R8 W8B581 A0A1B0AJS0 A0A0A1X8K9 A0A310SKX7 A0A034VW02 A0A0K8VIQ7 E0VVU9 A0A2P8XZJ7 A0A293MY73 A0A087SXH2 A0A1Z5L5A3 A0A1Z5KVR4 A0A1W4XUS7 U5EUZ1
Ontologies
GO
PANTHER
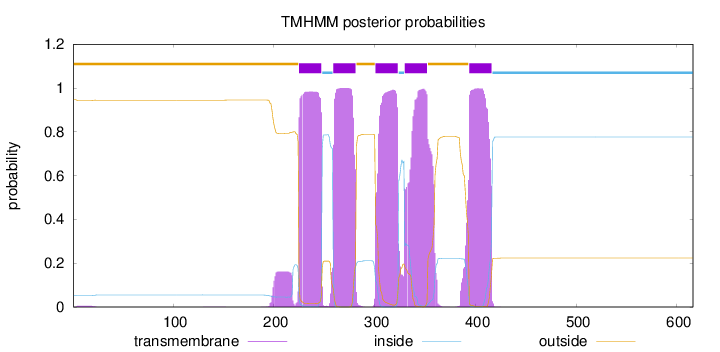
Topology
Length:
616
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
118.90225
Exp number, first 60 AAs:
0.09416
Total prob of N-in:
0.05269
outside
1 - 224
TMhelix
225 - 247
inside
248 - 258
TMhelix
259 - 281
outside
282 - 300
TMhelix
301 - 323
inside
324 - 329
TMhelix
330 - 352
outside
353 - 393
TMhelix
394 - 416
inside
417 - 616
Population Genetic Test Statistics
Pi
20.231142
Theta
19.931841
Tajima's D
-0.660357
CLR
484.027621
CSRT
0.207939603019849
Interpretation
Uncertain
