Gene
KWMTBOMO06543
Pre Gene Modal
BGIBMGA011831
Annotation
PREDICTED:_sodium/hydrogen_exchanger_3_[Bombyx_mori]
Full name
Sodium/hydrogen exchanger
Location in the cell
PlasmaMembrane Reliability : 4.502
Sequence
CDS
ATGACGTCATTGAAGCACGGAGTGAACTTGAGCAGACGCGCTTGCAGGCAAGCCTTCGCTTTGGTCGCTGTGAGCACGATACTGCACGCCGTACAATGTTCACCTGTCGAAACTACCACGAAAACGAATCCGTCCATGACAGTCGATGGTATATCGACGTACCCTGGTCTAGGGTTCGGACCAATGGGTCCTGTGGAGCCCTTACCCGGAGAGCCACCAGGCACACAGCTACGCCAGAACCAAGACACTACAACTGAATTACCGACAACTGAGAAAACCAAGAAGGAGAGGACCTTCGTGATAGCAGCGTTTGAGTTCCATCGAGTGGAGACGCCATTTTTGATCGGACTTTGGATATTTTTTGCCAGTCTCGCGAAGATAGGGTTCCACATGGCTCCGAAACTATCACGAATGTTACCTGAGTCATGTCTCCTTGTCTTCGTTGGTGTGTTGATAGGTGCATTACTTCTAAGTACTCACAGTGTACACGTTCAACCACTCACACCGGATACCTTCTTTTTGTATATGCTTCCACCTATAATATTGGATGCGGGATATTTTATGCCAAACAGGCTATTCTTTGATCATTTGGGTACCATTTTATTGTTTGCTGTTGTTGGGACTGTATTCAACACTTTGACTATTGGTGCTTCATTGTGGGCTTGCGGCCAGACTGGCATGTTCGGGTGTACAACACCGTTATTAGATATGCTGTTGTTCGGGGCTTTAATATCAGCCGTGGATCCGGTGGCGGTATTGGCGGTGTTCGAAGAAATTCACGTTGACGAAGTTTTGTATATTGTTGTTTTCGGTGAATCCTTACTTAACGATGCCGTTACTGTTGTATTGTATCACATGTTCGAAGCATACACGGAAATGGGACCGTCGCGTCTAGTCTACACGGATATTCTGGCCGGCCTGGCTTCGTTCCTGGTCGTAGCAGTAGGGGGAACGTGTATTGGAGTTGTGTGGGGCTTCGCGACGGGTCTCGTAACTCGCTTCACCAACGAAGTCAGGGTCATAGAGCCGATCTTCATATTCGTAATGGCGTATTTAGCATATTTGAACGCGGAAATGTTTCACATGAGCGGGATATTAGCCATAACATTTTGCGGTATTACAATGAAGAATTATGTTGAAGCGAATATTTCACATAAATCACACACCACAATCAAATACACTATGAAGATGCTATCGTCTTCGTCTGAGACCATTATATTCATGTTTCTCGGTGTCGCAACAGTGAATAACACCCACGTTTGGAATACGTGGTTCGTACTGTTAACCATTGTGTTCTGCTCAATATTCCGGATTATAGGAGTATACATATTTAGCGCTGTAGCAAATAAGTTTCGTCTGCACAAATTGAATAGAGTTGAGAAATTCGTCATGTCTTATGGAGGTCTCCGAGGCGCAGTGGCTTTCGCACTTGTTCTACTTATAGATCCAACGGTAGTAAATCTGCAGCCCATGTTTGTTACAACGACAATAGCTGTGATATACTTTACGGTTTTCATACAAGGCATCACGATCAAGCCCCTGGTCAAGATACTGAATGTGAAGACGGCGGAAAAACGGAAACCCACTATGAACGAGAGGATCCATGAGAGGTGCATGGATCACTTGATGGCAGGCATTGAAGATATTCTCGGCAAACATGGCAATCATCATATGCGTGATAAATTTAAAAGATTCGACAATAGATTTATCAGGCCATTCCTTTTGAGAAATTATCAGGGAGCGGAACCGAAGATTTTGGAGACTTATTCGAAATTAGCTATGAGAGACGCCATAGAATATATGCAAAGAAATGCTTCTACGATCGGTAACATTTCCGGAGCTGAATCCATGTCGGCGATTTTCAAAAATTACACAAACACAAACTTTAATGGAAGTCCAAGTTTTAGTCAAATAGATACATCGACTTGGAACATAGACATGCAGGAACTAGAATATAACCCCTCTAAGAAAGATCTCACTGATGCAAAAATCCATCATCTGCTCGCTGAAGAATTGTGCAAGCCTTTTAGACGGCATCGTCGCCTAAGCTACAGCCGGCATGCCGTAAACGAACGAGATCTCGGTATACAAGTCAACTACAAGACCCACATGAACATACGCAGACGGGTCGGTGACAGGAAACATCATCATAAAAGGAGCAAACGTGGAAAACAGGACGGAAAAAATCACGTTACTTTCCCGGAATTCCAGCAAAACGGTTCGGCTAAACAGTTCACGCACGATTATATCGACGAGGTACTTCAGGAGGACGGCGAAGAGCGGGAAGTCAGACGGGACGGCGACGACGGCGGTATTACATTCACTGCCAAATCCCCGCCCGGCTACAAAGAAGTCAGTCCAACGTCTTCGCATAAAGAAAATAGTAGTTCACTTTTAAACCAGTTGGCTAACCTGCCCGGATTTAATCCTCTTAAGGCGAGGATAATAAAGCAGTACGTTAAAACGCCCGAAGAGATACTACCGACAGCTGTTGAGAAATCCTTACCGTGGAGAAGGGACAGATCCACGTATAACTTTGTTCCCAACGAAGACATTCTAGAAGAAGACGAAAGAAGATTGTCCGATGACAACGATAACTACAACATGATCGTGGCTGAGCAAGCTCGAGTGGCGACGCCAACAGCAACGGAGACAATGTTGCCATGGAAACGAGAAGAAGCTGAAGGATCTGGTGCCCTCAAGCAATCAGAATTTCCTTCATGGGCTTCAAACAAAGAATATCTAGCCTATAATTCTCCTTCCTCTACTTTCTTAGGTCTATTCCGAAGAGAAAGCTCTAGTTCGACCCAAGGCGCTGCTGAAGCTGTAGTGCTATCTGATTCCGAGGGGCACAGAGAGAGTACCAGTAAAGGCGAAGCGTCCTCAAAAGACGGTGGTGTACGTCAAGGTCCTAGCTTATTATCGAAACGAAGCACAAGCCTGGGTGGTCCGTCTGTGTTGTTGATGGAGGACGTGGCCCATTCAGATCCTCTGGGTGCATCTAGCAGACCAGCTAGTATTATGCAAGGACCGCAACGAGGACAATGCCGGCGAGGTTCCATGATGGAGTTGACAGGTTCAATGTCGCGTGATGCAATAACAGAAGAGAAGTGTTGTAAATCGTTGCCAGAAAGTGAAAGGATGGGCGTCAGGCGTCTTCCCCTAGGTCAGAGTCTGGGACGGGAGCCGTTCATTCGACAGCTAACTTTAGCTTCGAGCCTTACGACTAGCTCATCATCAGAGGACTCATCGGACGAAAACACCTGA
Protein
MTSLKHGVNLSRRACRQAFALVAVSTILHAVQCSPVETTTKTNPSMTVDGISTYPGLGFGPMGPVEPLPGEPPGTQLRQNQDTTTELPTTEKTKKERTFVIAAFEFHRVETPFLIGLWIFFASLAKIGFHMAPKLSRMLPESCLLVFVGVLIGALLLSTHSVHVQPLTPDTFFLYMLPPIILDAGYFMPNRLFFDHLGTILLFAVVGTVFNTLTIGASLWACGQTGMFGCTTPLLDMLLFGALISAVDPVAVLAVFEEIHVDEVLYIVVFGESLLNDAVTVVLYHMFEAYTEMGPSRLVYTDILAGLASFLVVAVGGTCIGVVWGFATGLVTRFTNEVRVIEPIFIFVMAYLAYLNAEMFHMSGILAITFCGITMKNYVEANISHKSHTTIKYTMKMLSSSSETIIFMFLGVATVNNTHVWNTWFVLLTIVFCSIFRIIGVYIFSAVANKFRLHKLNRVEKFVMSYGGLRGAVAFALVLLIDPTVVNLQPMFVTTTIAVIYFTVFIQGITIKPLVKILNVKTAEKRKPTMNERIHERCMDHLMAGIEDILGKHGNHHMRDKFKRFDNRFIRPFLLRNYQGAEPKILETYSKLAMRDAIEYMQRNASTIGNISGAESMSAIFKNYTNTNFNGSPSFSQIDTSTWNIDMQELEYNPSKKDLTDAKIHHLLAEELCKPFRRHRRLSYSRHAVNERDLGIQVNYKTHMNIRRRVGDRKHHHKRSKRGKQDGKNHVTFPEFQQNGSAKQFTHDYIDEVLQEDGEEREVRRDGDDGGITFTAKSPPGYKEVSPTSSHKENSSSLLNQLANLPGFNPLKARIIKQYVKTPEEILPTAVEKSLPWRRDRSTYNFVPNEDILEEDERRLSDDNDNYNMIVAEQARVATPTATETMLPWKREEAEGSGALKQSEFPSWASNKEYLAYNSPSSTFLGLFRRESSSSTQGAAEAVVLSDSEGHRESTSKGEASSKDGGVRQGPSLLSKRSTSLGGPSVLLMEDVAHSDPLGASSRPASIMQGPQRGQCRRGSMMELTGSMSRDAITEEKCCKSLPESERMGVRRLPLGQSLGREPFIRQLTLASSLTTSSSSEDSSDENT
Summary
Similarity
Belongs to the monovalent cation:proton antiporter 1 (CPA1) transporter (TC 2.A.36) family.
Feature
chain Sodium/hydrogen exchanger
Uniprot
A0A2H1VK81
A0A212EQT5
A0A194PV44
A0A194QP30
A0A2A4JNX1
A0A2A4JNZ4
+ More
H9JQM5 A0A067R6I4 E0VVV0 A0A139WKN9 A0A139WKX4 A0A1S4H0P3 A0A139WKQ1 E2AT05 A0A310SUC0 A0A151X1K5 A0A158NEZ9 T1HLN6 A0A195EUA9 F4W456 A0A182Q8W0 A0A224XJ56 A0A1W6EVP8 A0A1I8JU83 A0A195CKH5 A0A1Y1M429 E2B6P9 A0A026WMV2 A0A1Y1M586 A0A154PCL4 A0A034VTN8 V9I7M5 A0A0L7QSZ7 W8AZ23 V9I6L7 A0A182IQG8 V9I6M0 A0A1Y1M1Z0 W8AV39 A0A087ZVR0 Q5TNB1 A0A084VYE0 V9I6L4 Q6Y8G3 A0A2M3ZYK3 A0A1I8Q7V1 B4KGP5 Q17L17 A0A1Q3G3S6 A0A1I8Q7Y4 Q7Z0L7 A0A195E5H0 A0A0Q9X5Y7 A0A1B6CUX3 Q9NCQ0 A0A1W4UHX1 A0A0C9PZD1 A0A0Q9X8H4 A0A2M4BBH5 A0A0Q9XFZ5 Q9VIF9 A0A2M3YX91 A0A0Q9X659 M9NDU7 A0A2M3ZF79 A0A182UBY0 A0A1W4UVN6 A0A1I8Q7U8 A0A1I8Q7W5 B4IFK5 B3NKU7 B4P6Z7 A0A1I8Q7Z7 B4MDP3 A0A1A9XW14 A0A0Q5VYS2 A0A1I8Q7U3 A0A1W4UX32 A0A0R1DNH7 A0A1Q3G3Q3 A0A1I8Q7W0 A0A1Q3G2W6 A0A0Q9WXB3 A0A1Q3G3K3 A0A0C9REV7 A0A1W4UHV5 A0A1A9ZK74 A0A2C9GPC7 A0A1Q3G3J1 A0A2M4CJH9 A0A2M4CJ94 B3MU54 A0A2M4CIY5 A0A0P8Y5P0 A0A1Q3G2Y7 A0A2M3YX89 A0A0Q9X5Z9 A0A2M3ZYE4
H9JQM5 A0A067R6I4 E0VVV0 A0A139WKN9 A0A139WKX4 A0A1S4H0P3 A0A139WKQ1 E2AT05 A0A310SUC0 A0A151X1K5 A0A158NEZ9 T1HLN6 A0A195EUA9 F4W456 A0A182Q8W0 A0A224XJ56 A0A1W6EVP8 A0A1I8JU83 A0A195CKH5 A0A1Y1M429 E2B6P9 A0A026WMV2 A0A1Y1M586 A0A154PCL4 A0A034VTN8 V9I7M5 A0A0L7QSZ7 W8AZ23 V9I6L7 A0A182IQG8 V9I6M0 A0A1Y1M1Z0 W8AV39 A0A087ZVR0 Q5TNB1 A0A084VYE0 V9I6L4 Q6Y8G3 A0A2M3ZYK3 A0A1I8Q7V1 B4KGP5 Q17L17 A0A1Q3G3S6 A0A1I8Q7Y4 Q7Z0L7 A0A195E5H0 A0A0Q9X5Y7 A0A1B6CUX3 Q9NCQ0 A0A1W4UHX1 A0A0C9PZD1 A0A0Q9X8H4 A0A2M4BBH5 A0A0Q9XFZ5 Q9VIF9 A0A2M3YX91 A0A0Q9X659 M9NDU7 A0A2M3ZF79 A0A182UBY0 A0A1W4UVN6 A0A1I8Q7U8 A0A1I8Q7W5 B4IFK5 B3NKU7 B4P6Z7 A0A1I8Q7Z7 B4MDP3 A0A1A9XW14 A0A0Q5VYS2 A0A1I8Q7U3 A0A1W4UX32 A0A0R1DNH7 A0A1Q3G3Q3 A0A1I8Q7W0 A0A1Q3G2W6 A0A0Q9WXB3 A0A1Q3G3K3 A0A0C9REV7 A0A1W4UHV5 A0A1A9ZK74 A0A2C9GPC7 A0A1Q3G3J1 A0A2M4CJH9 A0A2M4CJ94 B3MU54 A0A2M4CIY5 A0A0P8Y5P0 A0A1Q3G2Y7 A0A2M3YX89 A0A0Q9X5Z9 A0A2M3ZYE4
Pubmed
22118469
26354079
19121390
24845553
20566863
18362917
+ More
19820115 12364791 20798317 21347285 21719571 28004739 24508170 30249741 25348373 24495485 14747013 17210077 24438588 16943493 17994087 17510324 12386840 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 17550304
19820115 12364791 20798317 21347285 21719571 28004739 24508170 30249741 25348373 24495485 14747013 17210077 24438588 16943493 17994087 17510324 12386840 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 17550304
EMBL
ODYU01003010
SOQ41230.1
AGBW02013252
OWR43814.1
KQ459590
KPI97271.1
+ More
KQ461190 KPJ07272.1 NWSH01001010 PCG73143.1 PCG73142.1 BABH01012152 KK852854 KDR14983.1 DS235815 EEB17506.1 KQ971326 KYB28463.1 KYB28465.1 AAAB01008984 KYB28464.1 GL442439 EFN63440.1 KQ760208 OAD61382.1 KQ982585 KYQ54282.1 ADTU01001496 ADTU01001497 ADTU01001498 ADTU01001499 ACPB03012053 KQ981979 KYN31487.1 GL887491 EGI71052.1 AXCN02000627 GFTR01008365 JAW08061.1 KY563394 ARK19803.1 KQ977642 KYN00957.1 GEZM01043497 JAV79055.1 GL446000 EFN88633.1 KK107153 QOIP01000012 EZA56981.1 RLU15809.1 GEZM01043496 JAV79056.1 KQ434870 KZC09527.1 GAKP01013470 JAC45482.1 JR035716 JR035718 JR035721 JR035722 AEY56626.1 AEY56628.1 AEY56631.1 AEY56632.1 KQ414755 KOC61745.1 GAMC01016437 JAB90118.1 JR035709 JR035712 JR035715 JR035719 AEY56619.1 AEY56622.1 AEY56625.1 AEY56629.1 JR035711 JR035714 JR035717 JR035720 AEY56621.1 AEY56624.1 AEY56627.1 AEY56630.1 GEZM01043494 JAV79058.1 GAMC01016443 JAB90112.1 EAL39028.2 ATLV01018359 ATLV01018360 KE525231 KFB42984.1 JR035707 JR035708 JR035710 JR035713 AEY56617.1 AEY56618.1 AEY56620.1 AEY56623.1 AY170874 AAO34131.1 GGFK01000325 MBW33646.1 CH933807 EDW13245.2 CH477219 EAT47410.1 GFDL01000587 JAV34458.1 AY122621 AAM63432.1 KQ979609 KYN20147.1 KRG03638.1 GEDC01020016 JAS17282.1 AF187723 AAF80554.1 GBYB01006891 GBYB01006893 GBYB01006894 GBYB01006897 JAG76658.1 JAG76660.1 JAG76661.1 JAG76664.1 KRG03641.1 GGFJ01001263 MBW50404.1 KRG03646.1 AE014134 AAF53960.2 GGFM01000119 MBW20870.1 KRG03644.1 AFH03790.1 GGFM01006377 MBW27128.1 CH480833 EDW46427.1 CH954179 EDV54470.1 CM000158 EDW89966.1 CH940661 EDW71304.2 KQS61807.1 KRJ98837.1 GFDL01000615 JAV34430.1 GFDL01000902 JAV34143.1 KRF85646.1 GFDL01000658 JAV34387.1 GBYB01006895 JAG76662.1 APCN01005847 GFDL01000670 JAV34375.1 GGFL01001143 MBW65321.1 GGFL01001141 MBW65319.1 CH902624 EDV33383.1 GGFL01001142 MBW65320.1 KPU74447.1 GFDL01000882 JAV34163.1 GGFM01000120 MBW20871.1 KRG03654.1 GGFK01000253 MBW33574.1
KQ461190 KPJ07272.1 NWSH01001010 PCG73143.1 PCG73142.1 BABH01012152 KK852854 KDR14983.1 DS235815 EEB17506.1 KQ971326 KYB28463.1 KYB28465.1 AAAB01008984 KYB28464.1 GL442439 EFN63440.1 KQ760208 OAD61382.1 KQ982585 KYQ54282.1 ADTU01001496 ADTU01001497 ADTU01001498 ADTU01001499 ACPB03012053 KQ981979 KYN31487.1 GL887491 EGI71052.1 AXCN02000627 GFTR01008365 JAW08061.1 KY563394 ARK19803.1 KQ977642 KYN00957.1 GEZM01043497 JAV79055.1 GL446000 EFN88633.1 KK107153 QOIP01000012 EZA56981.1 RLU15809.1 GEZM01043496 JAV79056.1 KQ434870 KZC09527.1 GAKP01013470 JAC45482.1 JR035716 JR035718 JR035721 JR035722 AEY56626.1 AEY56628.1 AEY56631.1 AEY56632.1 KQ414755 KOC61745.1 GAMC01016437 JAB90118.1 JR035709 JR035712 JR035715 JR035719 AEY56619.1 AEY56622.1 AEY56625.1 AEY56629.1 JR035711 JR035714 JR035717 JR035720 AEY56621.1 AEY56624.1 AEY56627.1 AEY56630.1 GEZM01043494 JAV79058.1 GAMC01016443 JAB90112.1 EAL39028.2 ATLV01018359 ATLV01018360 KE525231 KFB42984.1 JR035707 JR035708 JR035710 JR035713 AEY56617.1 AEY56618.1 AEY56620.1 AEY56623.1 AY170874 AAO34131.1 GGFK01000325 MBW33646.1 CH933807 EDW13245.2 CH477219 EAT47410.1 GFDL01000587 JAV34458.1 AY122621 AAM63432.1 KQ979609 KYN20147.1 KRG03638.1 GEDC01020016 JAS17282.1 AF187723 AAF80554.1 GBYB01006891 GBYB01006893 GBYB01006894 GBYB01006897 JAG76658.1 JAG76660.1 JAG76661.1 JAG76664.1 KRG03641.1 GGFJ01001263 MBW50404.1 KRG03646.1 AE014134 AAF53960.2 GGFM01000119 MBW20870.1 KRG03644.1 AFH03790.1 GGFM01006377 MBW27128.1 CH480833 EDW46427.1 CH954179 EDV54470.1 CM000158 EDW89966.1 CH940661 EDW71304.2 KQS61807.1 KRJ98837.1 GFDL01000615 JAV34430.1 GFDL01000902 JAV34143.1 KRF85646.1 GFDL01000658 JAV34387.1 GBYB01006895 JAG76662.1 APCN01005847 GFDL01000670 JAV34375.1 GGFL01001143 MBW65321.1 GGFL01001141 MBW65319.1 CH902624 EDV33383.1 GGFL01001142 MBW65320.1 KPU74447.1 GFDL01000882 JAV34163.1 GGFM01000120 MBW20871.1 KRG03654.1 GGFK01000253 MBW33574.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000218220
UP000005204
UP000027135
+ More
UP000009046 UP000007266 UP000000311 UP000075809 UP000005205 UP000015103 UP000078541 UP000007755 UP000075886 UP000075900 UP000078542 UP000008237 UP000053097 UP000279307 UP000076502 UP000053825 UP000075880 UP000005203 UP000007062 UP000030765 UP000095300 UP000009192 UP000008820 UP000078492 UP000192221 UP000000803 UP000075902 UP000001292 UP000008711 UP000002282 UP000008792 UP000092443 UP000092445 UP000075840 UP000007801
UP000009046 UP000007266 UP000000311 UP000075809 UP000005205 UP000015103 UP000078541 UP000007755 UP000075886 UP000075900 UP000078542 UP000008237 UP000053097 UP000279307 UP000076502 UP000053825 UP000075880 UP000005203 UP000007062 UP000030765 UP000095300 UP000009192 UP000008820 UP000078492 UP000192221 UP000000803 UP000075902 UP000001292 UP000008711 UP000002282 UP000008792 UP000092443 UP000092445 UP000075840 UP000007801
Interpro
IPR018422
Cation/H_exchanger_CPA1
+ More
IPR006153 Cation/H_exchanger
IPR004709 NaH_exchanger
IPR018410 Na/H_exchanger_3/5
IPR036458 Na:dicarbo_symporter_sf
IPR001991 Na-dicarboxylate_symporter
IPR018107 Na-dicarboxylate_symporter_CS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR000719 Prot_kinase_dom
IPR020846 MFS_dom
IPR006153 Cation/H_exchanger
IPR004709 NaH_exchanger
IPR018410 Na/H_exchanger_3/5
IPR036458 Na:dicarbo_symporter_sf
IPR001991 Na-dicarboxylate_symporter
IPR018107 Na-dicarboxylate_symporter_CS
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR017441 Protein_kinase_ATP_BS
IPR000719 Prot_kinase_dom
IPR020846 MFS_dom
CDD
ProteinModelPortal
A0A2H1VK81
A0A212EQT5
A0A194PV44
A0A194QP30
A0A2A4JNX1
A0A2A4JNZ4
+ More
H9JQM5 A0A067R6I4 E0VVV0 A0A139WKN9 A0A139WKX4 A0A1S4H0P3 A0A139WKQ1 E2AT05 A0A310SUC0 A0A151X1K5 A0A158NEZ9 T1HLN6 A0A195EUA9 F4W456 A0A182Q8W0 A0A224XJ56 A0A1W6EVP8 A0A1I8JU83 A0A195CKH5 A0A1Y1M429 E2B6P9 A0A026WMV2 A0A1Y1M586 A0A154PCL4 A0A034VTN8 V9I7M5 A0A0L7QSZ7 W8AZ23 V9I6L7 A0A182IQG8 V9I6M0 A0A1Y1M1Z0 W8AV39 A0A087ZVR0 Q5TNB1 A0A084VYE0 V9I6L4 Q6Y8G3 A0A2M3ZYK3 A0A1I8Q7V1 B4KGP5 Q17L17 A0A1Q3G3S6 A0A1I8Q7Y4 Q7Z0L7 A0A195E5H0 A0A0Q9X5Y7 A0A1B6CUX3 Q9NCQ0 A0A1W4UHX1 A0A0C9PZD1 A0A0Q9X8H4 A0A2M4BBH5 A0A0Q9XFZ5 Q9VIF9 A0A2M3YX91 A0A0Q9X659 M9NDU7 A0A2M3ZF79 A0A182UBY0 A0A1W4UVN6 A0A1I8Q7U8 A0A1I8Q7W5 B4IFK5 B3NKU7 B4P6Z7 A0A1I8Q7Z7 B4MDP3 A0A1A9XW14 A0A0Q5VYS2 A0A1I8Q7U3 A0A1W4UX32 A0A0R1DNH7 A0A1Q3G3Q3 A0A1I8Q7W0 A0A1Q3G2W6 A0A0Q9WXB3 A0A1Q3G3K3 A0A0C9REV7 A0A1W4UHV5 A0A1A9ZK74 A0A2C9GPC7 A0A1Q3G3J1 A0A2M4CJH9 A0A2M4CJ94 B3MU54 A0A2M4CIY5 A0A0P8Y5P0 A0A1Q3G2Y7 A0A2M3YX89 A0A0Q9X5Z9 A0A2M3ZYE4
H9JQM5 A0A067R6I4 E0VVV0 A0A139WKN9 A0A139WKX4 A0A1S4H0P3 A0A139WKQ1 E2AT05 A0A310SUC0 A0A151X1K5 A0A158NEZ9 T1HLN6 A0A195EUA9 F4W456 A0A182Q8W0 A0A224XJ56 A0A1W6EVP8 A0A1I8JU83 A0A195CKH5 A0A1Y1M429 E2B6P9 A0A026WMV2 A0A1Y1M586 A0A154PCL4 A0A034VTN8 V9I7M5 A0A0L7QSZ7 W8AZ23 V9I6L7 A0A182IQG8 V9I6M0 A0A1Y1M1Z0 W8AV39 A0A087ZVR0 Q5TNB1 A0A084VYE0 V9I6L4 Q6Y8G3 A0A2M3ZYK3 A0A1I8Q7V1 B4KGP5 Q17L17 A0A1Q3G3S6 A0A1I8Q7Y4 Q7Z0L7 A0A195E5H0 A0A0Q9X5Y7 A0A1B6CUX3 Q9NCQ0 A0A1W4UHX1 A0A0C9PZD1 A0A0Q9X8H4 A0A2M4BBH5 A0A0Q9XFZ5 Q9VIF9 A0A2M3YX91 A0A0Q9X659 M9NDU7 A0A2M3ZF79 A0A182UBY0 A0A1W4UVN6 A0A1I8Q7U8 A0A1I8Q7W5 B4IFK5 B3NKU7 B4P6Z7 A0A1I8Q7Z7 B4MDP3 A0A1A9XW14 A0A0Q5VYS2 A0A1I8Q7U3 A0A1W4UX32 A0A0R1DNH7 A0A1Q3G3Q3 A0A1I8Q7W0 A0A1Q3G2W6 A0A0Q9WXB3 A0A1Q3G3K3 A0A0C9REV7 A0A1W4UHV5 A0A1A9ZK74 A0A2C9GPC7 A0A1Q3G3J1 A0A2M4CJH9 A0A2M4CJ94 B3MU54 A0A2M4CIY5 A0A0P8Y5P0 A0A1Q3G2Y7 A0A2M3YX89 A0A0Q9X5Z9 A0A2M3ZYE4
PDB
2MDF
E-value=1.43678e-16,
Score=215
Ontologies
GO
PANTHER
Topology
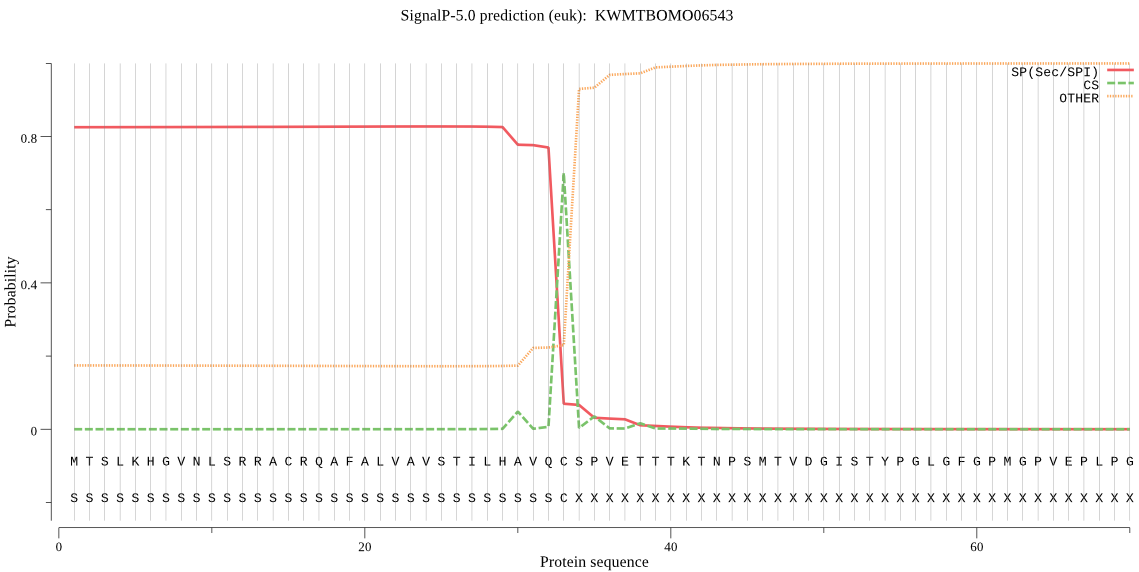
SignalP
Position: 1 - 33,
Likelihood: 0.828202
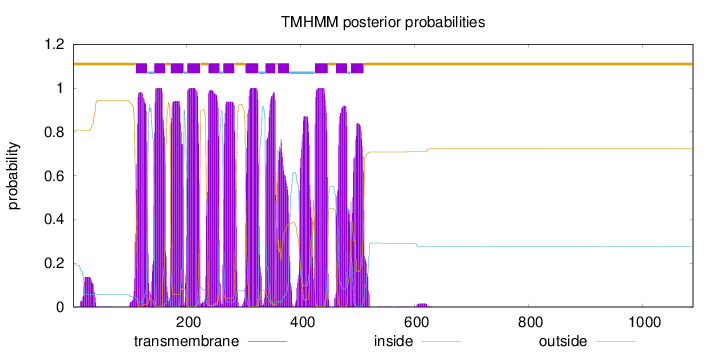
Length:
1088
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
260.888699999999
Exp number, first 60 AAs:
2.68255
Total prob of N-in:
0.19130
outside
1 - 110
TMhelix
111 - 130
inside
131 - 142
TMhelix
143 - 162
outside
163 - 171
TMhelix
172 - 194
inside
195 - 200
TMhelix
201 - 223
outside
224 - 237
TMhelix
238 - 257
inside
258 - 263
TMhelix
264 - 283
outside
284 - 302
TMhelix
303 - 325
inside
326 - 337
TMhelix
338 - 355
outside
356 - 359
TMhelix
360 - 379
inside
380 - 424
TMhelix
425 - 447
outside
448 - 461
TMhelix
462 - 481
inside
482 - 487
TMhelix
488 - 510
outside
511 - 1088
Population Genetic Test Statistics
Pi
227.239083
Theta
184.644448
Tajima's D
0.818786
CLR
0.293288
CSRT
0.605969701514924
Interpretation
Uncertain
