Gene
KWMTBOMO06541
Pre Gene Modal
BGIBMGA011834
Annotation
methionine_sulfoxide_reductase_[Bombyx_mori]
Full name
Peptide methionine sulfoxide reductase
Alternative Name
Ecdysone-induced protein 28/29 kDa
Methionine-S-sulfoxide reductase
Peptide-methionine (S)-S-oxide reductase
Methionine-S-sulfoxide reductase
Peptide-methionine (S)-S-oxide reductase
Location in the cell
Cytoplasmic Reliability : 1.169 Mitochondrial Reliability : 1.651
Sequence
CDS
ATGAAGCCACCAGTTAACAACACCCTTTTGCACGATGTGGACATTCCCACGAAGAAGGCGACTTTTGGTATGGGCTGCTTCTGGTCAAACGATTCTCTTTTCGGCGCCACGCTCGGTGTCATACGCACTCGTGTGGGATACGCCGGTGGCACGACCAAGAATCCGCATTACAGAAGCATCGGGGACCACACAGAAGTTATAGAGATCGATTATGATCCGAAAACTGTTACCTATGAGGAGCTTCTCGAAATTTTCTGGGCTAACCACGAGTACGGACTAACCACCAAATTAAAGAGACAGTATCAGTCAATGATTCTGTACCACGATGATGAACAACGTTTAGTGGCTGAAGCCTCGCACAGAGCCGTGCAGAGCCGCTGTAACGAGACCTTACGCACTGAGATAGCGAAGGCTGACGTTTTCTATCCAGCTGAAGACTACCATCAGAAATACAGACTTCAAGGCCACAAAGACTTCTGCCGCAGTATCGGGCTCGATTCCTCTAAGCTACAGACTTCTCATTTGGCCGCCAGACTCAACGGTTACCTGGTCGGTGTTGGAGGAATGAATCAATTCGACCAGGAACTGCCGAAACTAGGCCTCAACGAGAAACAAGCTGAATACGTGAGGAGGGAGCTTGAACGCAACGAAGGCGGCGGTCTGGCTTGCTAA
Protein
MKPPVNNTLLHDVDIPTKKATFGMGCFWSNDSLFGATLGVIRTRVGYAGGTTKNPHYRSIGDHTEVIEIDYDPKTVTYEELLEIFWANHEYGLTTKLKRQYQSMILYHDDEQRLVAEASHRAVQSRCNETLRTEIAKADVFYPAEDYHQKYRLQGHKDFCRSIGLDSSKLQTSHLAARLNGYLVGVGGMNQFDQELPKLGLNEKQAEYVRRELERNEGGGLAC
Summary
Description
Has an important function as a repair enzyme for proteins that have been inactivated by oxidation. Catalyzes the reversible oxidation-reduction of methionine sulfoxide in proteins to methionine.
Catalytic Activity
[thioredoxin]-disulfide + H2O + L-methionyl-[protein] = [thioredoxin]-dithiol + L-methionyl-(S)-S-oxide-[protein]
[thioredoxin]-disulfide + H2O + L-methionine = [thioredoxin]-dithiol + L-methionine (S)-S-oxide
[thioredoxin]-disulfide + H2O + L-methionine = [thioredoxin]-dithiol + L-methionine (S)-S-oxide
Similarity
Belongs to the MsrA Met sulfoxide reductase family.
Keywords
Alternative splicing
Complete proteome
Oxidoreductase
Reference proteome
Ubl conjugation
Feature
chain Peptide methionine sulfoxide reductase
splice variant In isoform Eip29.
splice variant In isoform Eip29.
Uniprot
Q1HQ83
H9JQM8
S4PTD2
A0A2A4IZQ8
A0A212EQP4
I4DKF1
+ More
A0A2H1VK86 A0A3S2LZV9 A0A194PWY0 I4DS31 A0A0L7LBI5 A0A084WIV9 A0A182J992 A0A1B0CT88 A0A1L8DU60 A0A2M3YZ62 A0A2C9GQY1 Q7PVA0 A0A182Q1P8 A0A2M4BZV0 A0A182KSB4 A0A2M4BYD7 Q7Q0D3 Q1HRN8 D6WWX6 Q17NK7 A0A023EKL0 A0A2M4CUY5 A0A182TB04 A0A0K8TLT4 A0A2M4A5T9 A0A182Y1I0 V5H0U7 A0A182FBD2 A0A1Q3FKK8 A0A1J1J3M2 B0XGG5 A0A182P5W1 A0A1L8EEL7 A0A0L0C8I9 A0A1L8EEI9 U5ESG1 A0A1L8EEP9 A0A1L8EJ87 A0A1L8EEL4 A0A1L8EEW3 A0A1L8EEK2 A0A1L8E9B6 A0A1L8E9M4 A0A1Q3FJ24 A0A1L8EAE8 A0A034VIH8 A0A1W4VPE2 A0A182GKA0 A0A1W4VNS0 A0A1W4VB12 W8BSG7 Q17NK6 A0A194QPJ4 W8C9F0 A0A1I8MSM3 A0A1I8MSM1 Q6NLA4 B0XGG4 B0WFN8 A0A1I8MSP1 A0A182KSA9 D3TL12 A0A182P5W2 A0A1B0F9B2 A0A1A9ZDP0 A0A0A1X6Q3 B4HI85 B4QL24 A0A1A9Y1K7 A0A1B0BPV4 A0A1A9UM08 B5X0L2 P08761 A0A0J9RW08 A0A0R1E8A1 B3NI28 Q7KUM8 A0A1I8PSG9 A0A1I8PSH0 A0A0R1ECT8 A0A0J9ULI0 A0A1I8PSD3 B4NLV9 M9PI66 M9PCL6 A0A1I8PSH7 B4GUB8 A0A1Y1L9S4 A0A0R3P7U0 A0A1Y1LAT4 B3M707 A0A1I8PSD2 A0A026W992 A0A1A9W4D9
A0A2H1VK86 A0A3S2LZV9 A0A194PWY0 I4DS31 A0A0L7LBI5 A0A084WIV9 A0A182J992 A0A1B0CT88 A0A1L8DU60 A0A2M3YZ62 A0A2C9GQY1 Q7PVA0 A0A182Q1P8 A0A2M4BZV0 A0A182KSB4 A0A2M4BYD7 Q7Q0D3 Q1HRN8 D6WWX6 Q17NK7 A0A023EKL0 A0A2M4CUY5 A0A182TB04 A0A0K8TLT4 A0A2M4A5T9 A0A182Y1I0 V5H0U7 A0A182FBD2 A0A1Q3FKK8 A0A1J1J3M2 B0XGG5 A0A182P5W1 A0A1L8EEL7 A0A0L0C8I9 A0A1L8EEI9 U5ESG1 A0A1L8EEP9 A0A1L8EJ87 A0A1L8EEL4 A0A1L8EEW3 A0A1L8EEK2 A0A1L8E9B6 A0A1L8E9M4 A0A1Q3FJ24 A0A1L8EAE8 A0A034VIH8 A0A1W4VPE2 A0A182GKA0 A0A1W4VNS0 A0A1W4VB12 W8BSG7 Q17NK6 A0A194QPJ4 W8C9F0 A0A1I8MSM3 A0A1I8MSM1 Q6NLA4 B0XGG4 B0WFN8 A0A1I8MSP1 A0A182KSA9 D3TL12 A0A182P5W2 A0A1B0F9B2 A0A1A9ZDP0 A0A0A1X6Q3 B4HI85 B4QL24 A0A1A9Y1K7 A0A1B0BPV4 A0A1A9UM08 B5X0L2 P08761 A0A0J9RW08 A0A0R1E8A1 B3NI28 Q7KUM8 A0A1I8PSG9 A0A1I8PSH0 A0A0R1ECT8 A0A0J9ULI0 A0A1I8PSD3 B4NLV9 M9PI66 M9PCL6 A0A1I8PSH7 B4GUB8 A0A1Y1L9S4 A0A0R3P7U0 A0A1Y1LAT4 B3M707 A0A1I8PSD2 A0A026W992 A0A1A9W4D9
EC Number
1.8.4.11
Pubmed
19121390
23622113
22118469
22651552
26354079
26227816
+ More
24438588 12364791 14747013 17210077 20966253 17204158 18362917 19820115 17510324 24945155 26483478 26369729 25244985 26108605 25348373 24495485 25315136 20353571 25830018 17994087 22936249 3097323 12145281 10731132 12537572 28953965 17550304 12537568 12537573 12537574 16110336 17569856 17569867 28004739 15632085 24508170 30249741
24438588 12364791 14747013 17210077 20966253 17204158 18362917 19820115 17510324 24945155 26483478 26369729 25244985 26108605 25348373 24495485 25315136 20353571 25830018 17994087 22936249 3097323 12145281 10731132 12537572 28953965 17550304 12537568 12537573 12537574 16110336 17569856 17569867 28004739 15632085 24508170 30249741
EMBL
DQ443169
ABF51258.1
BABH01012156
GAIX01012243
JAA80317.1
NWSH01004534
+ More
PCG64976.1 AGBW02013253 OWR43812.1 AK401769 BAM18391.1 ODYU01003010 SOQ41233.1 RSAL01000094 RVE47866.1 KQ459590 KPI97269.1 AK405394 BAM20721.1 JTDY01001838 KOB72754.1 ATLV01023950 KE525347 KFB50153.1 AJWK01027294 GFDF01004134 JAV09950.1 GGFM01000802 MBW21553.1 APCN01003307 AAAB01008986 EAA00184.4 AXCN02001024 GGFJ01009270 MBW58411.1 GGFJ01008956 MBW58097.1 EAA00212.3 DQ440056 ABF18089.1 KQ971361 EFA08080.1 CH477198 EAT48308.1 JXUM01069649 GAPW01003903 GAPW01003902 KQ562562 JAC09696.1 KXJ75615.1 GGFL01004921 MBW69099.1 GDAI01002505 JAI15098.1 GGFK01002846 MBW36167.1 GALX01002013 JAB66453.1 GFDL01006905 JAV28140.1 CVRI01000069 CRL07045.1 DS233029 EDS27596.1 GFDG01001668 JAV17131.1 JRES01000760 KNC28560.1 GFDG01001667 JAV17132.1 GANO01002372 JAB57499.1 GFDG01001674 JAV17125.1 GFDG01000110 JAV18689.1 GFDG01001671 JAV17128.1 GFDG01001670 JAV17129.1 GFDG01001669 JAV17130.1 GFDG01003515 JAV15284.1 GFDG01003514 JAV15285.1 GFDL01007435 JAV27610.1 GFDG01003224 JAV15575.1 GAKP01015856 JAC43096.1 KXJ75616.1 GAMC01006707 JAB99848.1 EAT48309.1 KQ461190 KPJ07274.1 GAMC01006708 GAMC01006706 JAB99849.1 BT012430 AAS93701.1 EDS27595.1 DS231920 EDS26382.1 EZ422103 ADD18307.1 CCAG010023097 GBXI01007974 JAD06318.1 CH480815 EDW41581.1 CM000363 CM002912 EDX10549.1 KMY99748.1 JXJN01018237 BT044581 ACI16543.1 X58286 X04024 X04521 AF541958 AE014296 BT004857 KMY99752.1 CH891752 KRK05263.1 CH954178 EDV52114.2 AAS64994.2 KRK05265.1 KMY99750.1 CH964278 EDW85411.1 AGB94571.1 AGB94572.1 CH479191 EDW26201.1 GEZM01064621 JAV68685.1 CH379069 KRT07504.1 GEZM01064620 JAV68686.1 CH902618 EDV40872.2 KK107321 QOIP01000002 EZA52657.1 RLU25906.1
PCG64976.1 AGBW02013253 OWR43812.1 AK401769 BAM18391.1 ODYU01003010 SOQ41233.1 RSAL01000094 RVE47866.1 KQ459590 KPI97269.1 AK405394 BAM20721.1 JTDY01001838 KOB72754.1 ATLV01023950 KE525347 KFB50153.1 AJWK01027294 GFDF01004134 JAV09950.1 GGFM01000802 MBW21553.1 APCN01003307 AAAB01008986 EAA00184.4 AXCN02001024 GGFJ01009270 MBW58411.1 GGFJ01008956 MBW58097.1 EAA00212.3 DQ440056 ABF18089.1 KQ971361 EFA08080.1 CH477198 EAT48308.1 JXUM01069649 GAPW01003903 GAPW01003902 KQ562562 JAC09696.1 KXJ75615.1 GGFL01004921 MBW69099.1 GDAI01002505 JAI15098.1 GGFK01002846 MBW36167.1 GALX01002013 JAB66453.1 GFDL01006905 JAV28140.1 CVRI01000069 CRL07045.1 DS233029 EDS27596.1 GFDG01001668 JAV17131.1 JRES01000760 KNC28560.1 GFDG01001667 JAV17132.1 GANO01002372 JAB57499.1 GFDG01001674 JAV17125.1 GFDG01000110 JAV18689.1 GFDG01001671 JAV17128.1 GFDG01001670 JAV17129.1 GFDG01001669 JAV17130.1 GFDG01003515 JAV15284.1 GFDG01003514 JAV15285.1 GFDL01007435 JAV27610.1 GFDG01003224 JAV15575.1 GAKP01015856 JAC43096.1 KXJ75616.1 GAMC01006707 JAB99848.1 EAT48309.1 KQ461190 KPJ07274.1 GAMC01006708 GAMC01006706 JAB99849.1 BT012430 AAS93701.1 EDS27595.1 DS231920 EDS26382.1 EZ422103 ADD18307.1 CCAG010023097 GBXI01007974 JAD06318.1 CH480815 EDW41581.1 CM000363 CM002912 EDX10549.1 KMY99748.1 JXJN01018237 BT044581 ACI16543.1 X58286 X04024 X04521 AF541958 AE014296 BT004857 KMY99752.1 CH891752 KRK05263.1 CH954178 EDV52114.2 AAS64994.2 KRK05265.1 KMY99750.1 CH964278 EDW85411.1 AGB94571.1 AGB94572.1 CH479191 EDW26201.1 GEZM01064621 JAV68685.1 CH379069 KRT07504.1 GEZM01064620 JAV68686.1 CH902618 EDV40872.2 KK107321 QOIP01000002 EZA52657.1 RLU25906.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053268
UP000037510
+ More
UP000030765 UP000075880 UP000092461 UP000075840 UP000007062 UP000075886 UP000075882 UP000007266 UP000008820 UP000069940 UP000249989 UP000075901 UP000076408 UP000069272 UP000183832 UP000002320 UP000075885 UP000037069 UP000192221 UP000053240 UP000095301 UP000092444 UP000092445 UP000001292 UP000000304 UP000092443 UP000092460 UP000078200 UP000000803 UP000002282 UP000008711 UP000095300 UP000007798 UP000008744 UP000001819 UP000007801 UP000053097 UP000279307 UP000091820
UP000030765 UP000075880 UP000092461 UP000075840 UP000007062 UP000075886 UP000075882 UP000007266 UP000008820 UP000069940 UP000249989 UP000075901 UP000076408 UP000069272 UP000183832 UP000002320 UP000075885 UP000037069 UP000192221 UP000053240 UP000095301 UP000092444 UP000092445 UP000001292 UP000000304 UP000092443 UP000092460 UP000078200 UP000000803 UP000002282 UP000008711 UP000095300 UP000007798 UP000008744 UP000001819 UP000007801 UP000053097 UP000279307 UP000091820
Interpro
Gene 3D
CDD
ProteinModelPortal
Q1HQ83
H9JQM8
S4PTD2
A0A2A4IZQ8
A0A212EQP4
I4DKF1
+ More
A0A2H1VK86 A0A3S2LZV9 A0A194PWY0 I4DS31 A0A0L7LBI5 A0A084WIV9 A0A182J992 A0A1B0CT88 A0A1L8DU60 A0A2M3YZ62 A0A2C9GQY1 Q7PVA0 A0A182Q1P8 A0A2M4BZV0 A0A182KSB4 A0A2M4BYD7 Q7Q0D3 Q1HRN8 D6WWX6 Q17NK7 A0A023EKL0 A0A2M4CUY5 A0A182TB04 A0A0K8TLT4 A0A2M4A5T9 A0A182Y1I0 V5H0U7 A0A182FBD2 A0A1Q3FKK8 A0A1J1J3M2 B0XGG5 A0A182P5W1 A0A1L8EEL7 A0A0L0C8I9 A0A1L8EEI9 U5ESG1 A0A1L8EEP9 A0A1L8EJ87 A0A1L8EEL4 A0A1L8EEW3 A0A1L8EEK2 A0A1L8E9B6 A0A1L8E9M4 A0A1Q3FJ24 A0A1L8EAE8 A0A034VIH8 A0A1W4VPE2 A0A182GKA0 A0A1W4VNS0 A0A1W4VB12 W8BSG7 Q17NK6 A0A194QPJ4 W8C9F0 A0A1I8MSM3 A0A1I8MSM1 Q6NLA4 B0XGG4 B0WFN8 A0A1I8MSP1 A0A182KSA9 D3TL12 A0A182P5W2 A0A1B0F9B2 A0A1A9ZDP0 A0A0A1X6Q3 B4HI85 B4QL24 A0A1A9Y1K7 A0A1B0BPV4 A0A1A9UM08 B5X0L2 P08761 A0A0J9RW08 A0A0R1E8A1 B3NI28 Q7KUM8 A0A1I8PSG9 A0A1I8PSH0 A0A0R1ECT8 A0A0J9ULI0 A0A1I8PSD3 B4NLV9 M9PI66 M9PCL6 A0A1I8PSH7 B4GUB8 A0A1Y1L9S4 A0A0R3P7U0 A0A1Y1LAT4 B3M707 A0A1I8PSD2 A0A026W992 A0A1A9W4D9
A0A2H1VK86 A0A3S2LZV9 A0A194PWY0 I4DS31 A0A0L7LBI5 A0A084WIV9 A0A182J992 A0A1B0CT88 A0A1L8DU60 A0A2M3YZ62 A0A2C9GQY1 Q7PVA0 A0A182Q1P8 A0A2M4BZV0 A0A182KSB4 A0A2M4BYD7 Q7Q0D3 Q1HRN8 D6WWX6 Q17NK7 A0A023EKL0 A0A2M4CUY5 A0A182TB04 A0A0K8TLT4 A0A2M4A5T9 A0A182Y1I0 V5H0U7 A0A182FBD2 A0A1Q3FKK8 A0A1J1J3M2 B0XGG5 A0A182P5W1 A0A1L8EEL7 A0A0L0C8I9 A0A1L8EEI9 U5ESG1 A0A1L8EEP9 A0A1L8EJ87 A0A1L8EEL4 A0A1L8EEW3 A0A1L8EEK2 A0A1L8E9B6 A0A1L8E9M4 A0A1Q3FJ24 A0A1L8EAE8 A0A034VIH8 A0A1W4VPE2 A0A182GKA0 A0A1W4VNS0 A0A1W4VB12 W8BSG7 Q17NK6 A0A194QPJ4 W8C9F0 A0A1I8MSM3 A0A1I8MSM1 Q6NLA4 B0XGG4 B0WFN8 A0A1I8MSP1 A0A182KSA9 D3TL12 A0A182P5W2 A0A1B0F9B2 A0A1A9ZDP0 A0A0A1X6Q3 B4HI85 B4QL24 A0A1A9Y1K7 A0A1B0BPV4 A0A1A9UM08 B5X0L2 P08761 A0A0J9RW08 A0A0R1E8A1 B3NI28 Q7KUM8 A0A1I8PSG9 A0A1I8PSH0 A0A0R1ECT8 A0A0J9ULI0 A0A1I8PSD3 B4NLV9 M9PI66 M9PCL6 A0A1I8PSH7 B4GUB8 A0A1Y1L9S4 A0A0R3P7U0 A0A1Y1LAT4 B3M707 A0A1I8PSD2 A0A026W992 A0A1A9W4D9
PDB
4U66
E-value=1.16474e-36,
Score=381
Ontologies
GO
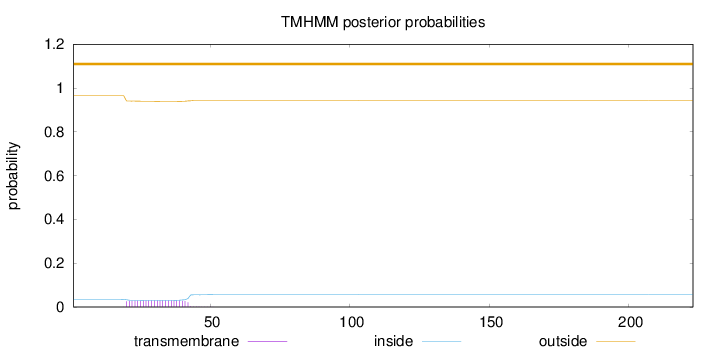
Topology
Length:
223
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.686700000000001
Exp number, first 60 AAs:
0.68465
Total prob of N-in:
0.03465
outside
1 - 223
Population Genetic Test Statistics
Pi
153.6538
Theta
136.844355
Tajima's D
-1.047374
CLR
114.829506
CSRT
0.124443777811109
Interpretation
Possibly Positive selection
