Gene
KWMTBOMO06538 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011835
Annotation
thioesterase-Ap_[Antheraea_pernyi]
Location in the cell
Extracellular Reliability : 1.621
Sequence
CDS
ATGAGGTTGTGGTTAATTCCATTGTTATTAATAAAATTAATAAGCGCAACGCCTACACCCATCGTCTTGTGGCACGGCATGGGTGACACATGCTGCATGTCCTTTAGTCTTGGTTCCTTCAAAATATTCCTAGAGAAGAACATACCTGGTGTTTATGTTCTGTCTCTCCAAATTGGCAATAATACTGTTGAAGATTTTGAAAATGGATATTTCATGAACCCGAATCTCCAAGTTGAATATGTATGTGAGAAGTTAGCGGCCGATCCAAAACTATCAAACGGATTCAATGTCATGGGTTTCTCACAGGGATGTCAGTTTATTCGAGCTGTAGTACAAAGATGTGGTCATAAACTACCACAAATTAAAAACATGGTGTCTCTCGGAGGTCAGCACCAAGGCATCTATGGCATACCTCATTGTGGTGCCTTGAGACACGAAACATGTGATGATGTGAGGAAGCTACTCAATTACGCTGCATATAACAGCTGGGTACAAAATTCACTGGTTCAAGCGACATATTGGCACGATCCTTTAGACGAGCGGACATATGAGGCAAACAGCGTATTTCTAGCGGACATCAACAACGTGAGGACCGTCAACAAGACGTATATACAGAACTTGAATAATCTAGAAAGATTCGTGCTGGTGATGTTCGATAATGATAGCATAGTGCAGCCGAAGGAGACGGAATGGTTCGGTTTTTACGCTCCGGGACAGGCCGAGAAATTGGTATCGTTAAGGGAATCTAAGATTTATTTGCAGGATCGACTCGGCCTCAAGAAAATGGATAAAGCGGGAAAATTAATTTTCTTATCAACGCCTGGCGACCATCTACGTTTCACAGAAGAATGGATGATCAAAAACATAATTAAACCTTATCTTTCTAGTTAA
Protein
MRLWLIPLLLIKLISATPTPIVLWHGMGDTCCMSFSLGSFKIFLEKNIPGVYVLSLQIGNNTVEDFENGYFMNPNLQVEYVCEKLAADPKLSNGFNVMGFSQGCQFIRAVVQRCGHKLPQIKNMVSLGGQHQGIYGIPHCGALRHETCDDVRKLLNYAAYNSWVQNSLVQATYWHDPLDERTYEANSVFLADINNVRTVNKTYIQNLNNLERFVLVMFDNDSIVQPKETEWFGFYAPGQAEKLVSLRESKIYLQDRLGLKKMDKAGKLIFLSTPGDHLRFTEEWMIKNIIKPYLSS
Summary
Uniprot
H9JQM9
W5VH72
D2SNX5
A0A2A4K346
A0A291P140
A0A194PVB8
+ More
A0A291P109 I4DJL9 A0A1V0M8B3 A0A088M9W1 A0A2H1VVX5 A0A2W1BQ24 A0A212FIN8 A0A1E1WC38 U4UAS9 J3JUY4 A0A1B6M6X5 A0A232EH28 K7IXU0 A0A267DQY9 A0A1Y1MLU8 A0A2J7QLI3 A0A1I8HQQ4 A0A1I8HWS3 A0A1B6H0L4 A0A067RBT8 A0A1L8DPX5 A0A1L8DPR4 A0A1L8DPR5 A0A1L8DPZ2 D6W6C0 H3B5N4 A0A336MSF6 C3ZCW0 A0A1Q3FJJ0 A0A182NDS4 B0W1X8 F6USM5 Q6DEU7 A2VDB5 A0A1L8HFT8 A0A1S4G3D1 Q170Q3 A0A1S4FHM6 A0A3B3BHH4 A7S9F2 Q16ET0 Q5WPS7 T1DPS5 R7VM55 A0A182PDH7 A0A1Y9J0D5 E9IAX5 W5LXU3 A0A250YFA7 Q7QDD6 A0A1W4XJH9 A0A1U7V115 S4RY76 A0A182IEP5 A0A3P8VM80 I3M5E3 T2FF50 A0A0P4WEX6 A0A0T6B4G0 A0A3B4BKW8 A0A182UVS8 A0A182YEM3 A0A1B6BZS0 A0A146M132 A0A023EP03 A0A146LCT3 A0A182GFT6 W8C624 B4M1A7 M3YS55 G9KIB9 A0A0A9Y3V5 A0A287AYH9 A0A146L3N2 G1LCS6 A0A336LF62 A0A2Y9IUU8 A0A384DT43 A0A3Q7XYH0 D2HXM1 G3NDR8 A0A0A9WSG7 A1L2U1 A0A1B0D0F5 A0A3Q2UST9 T1PCN1 A0A3P8U2Q8 Q5JZR0 A0A2U3X096 A0A1I8NAF5 A0A310UCB0 A0A3Q4HLX5 G3TPC5 A0A2M4BVN7
A0A291P109 I4DJL9 A0A1V0M8B3 A0A088M9W1 A0A2H1VVX5 A0A2W1BQ24 A0A212FIN8 A0A1E1WC38 U4UAS9 J3JUY4 A0A1B6M6X5 A0A232EH28 K7IXU0 A0A267DQY9 A0A1Y1MLU8 A0A2J7QLI3 A0A1I8HQQ4 A0A1I8HWS3 A0A1B6H0L4 A0A067RBT8 A0A1L8DPX5 A0A1L8DPR4 A0A1L8DPR5 A0A1L8DPZ2 D6W6C0 H3B5N4 A0A336MSF6 C3ZCW0 A0A1Q3FJJ0 A0A182NDS4 B0W1X8 F6USM5 Q6DEU7 A2VDB5 A0A1L8HFT8 A0A1S4G3D1 Q170Q3 A0A1S4FHM6 A0A3B3BHH4 A7S9F2 Q16ET0 Q5WPS7 T1DPS5 R7VM55 A0A182PDH7 A0A1Y9J0D5 E9IAX5 W5LXU3 A0A250YFA7 Q7QDD6 A0A1W4XJH9 A0A1U7V115 S4RY76 A0A182IEP5 A0A3P8VM80 I3M5E3 T2FF50 A0A0P4WEX6 A0A0T6B4G0 A0A3B4BKW8 A0A182UVS8 A0A182YEM3 A0A1B6BZS0 A0A146M132 A0A023EP03 A0A146LCT3 A0A182GFT6 W8C624 B4M1A7 M3YS55 G9KIB9 A0A0A9Y3V5 A0A287AYH9 A0A146L3N2 G1LCS6 A0A336LF62 A0A2Y9IUU8 A0A384DT43 A0A3Q7XYH0 D2HXM1 G3NDR8 A0A0A9WSG7 A1L2U1 A0A1B0D0F5 A0A3Q2UST9 T1PCN1 A0A3P8U2Q8 Q5JZR0 A0A2U3X096 A0A1I8NAF5 A0A310UCB0 A0A3Q4HLX5 G3TPC5 A0A2M4BVN7
Pubmed
19121390
20074338
28986331
26354079
22651552
28349297
+ More
26385554 28756777 22118469 23537049 22516182 28648823 20075255 28004739 26392545 24845553 18362917 19820115 9215903 18563158 20431018 27762356 17510324 29451363 17615350 15371479 23254933 21282665 28087693 12364791 14747013 17210077 24487278 25463417 25244985 26823975 24945155 26483478 24495485 17994087 23236062 25401762 30723633 20010809 24813606 16341006 25315136 25186727
26385554 28756777 22118469 23537049 22516182 28648823 20075255 28004739 26392545 24845553 18362917 19820115 9215903 18563158 20431018 27762356 17510324 29451363 17615350 15371479 23254933 21282665 28087693 12364791 14747013 17210077 24487278 25463417 25244985 26823975 24945155 26483478 24495485 17994087 23236062 25401762 30723633 20010809 24813606 16341006 25315136 25186727
EMBL
BABH01012161
KF670140
AHH80648.1
EZ407260
ACX53816.1
NWSH01000214
+ More
PCG78348.1 MF706190 ATJ44617.1 KQ459590 KPI97267.1 MF687646 ATJ44572.1 AK401487 BAM18109.1 KU755516 ARD71226.1 KJ579215 AIN34691.1 ODYU01004769 SOQ45001.1 KZ149984 PZC75714.1 AGBW02008350 OWR53605.1 GDQN01006492 JAT84562.1 KB632168 ERL89413.1 BT127049 AEE62011.1 GEBQ01008324 JAT31653.1 NNAY01004605 OXU17666.1 NIVC01003373 PAA51713.1 GEZM01029597 JAV86058.1 NEVH01013243 PNF29443.1 NIVC01001242 PAA70398.1 NIVC01001300 PAA69680.1 GECZ01016308 GECZ01005157 GECZ01001578 GECZ01000089 JAS53461.1 JAS64612.1 JAS68191.1 JAS69680.1 KK852567 KDR21202.1 GFDF01005633 JAV08451.1 GFDF01005632 JAV08452.1 GFDF01005638 JAV08446.1 GFDF01005637 JAV08447.1 KQ971319 EFA11379.1 AFYH01052445 AFYH01052446 UFQS01002445 UFQT01002445 SSX14055.1 SSX33474.1 GG666610 EEN49613.1 GFDL01007254 JAV27791.1 DS231824 EDS27414.1 AAMC01027184 AAMC01027185 BC076997 CR848336 AAH76997.1 CAJ83332.1 BC129713 AAI29714.1 CM004468 OCT94947.1 CH477470 EAT40438.1 DS469603 EDO39662.1 CH478592 EAT32741.1 AY455918 AAS16918.1 GAMD01001680 JAA99910.1 AMQN01003821 AMQN01003822 KB292015 ELU18250.1 GL762091 EFZ22264.1 AHAT01009296 GFFW01002564 JAV42224.1 AAAB01008852 EAA07412.4 APCN01009249 AGTP01008444 AGTP01008445 KF143809 AGV76999.1 GDRN01075798 JAI62990.1 LJIG01009943 KRT82069.1 GEDC01030531 JAS06767.1 GDHC01020622 GDHC01005136 JAP98006.1 JAQ13493.1 JXUM01046406 GAPW01002907 KQ561475 JAC10691.1 KXJ78478.1 GDHC01013294 JAQ05335.1 JXUM01060399 KQ562100 KXJ76693.1 GAMC01004539 JAC02017.1 CH940651 EDW65461.2 AEYP01007624 AEYP01007625 JP016046 AES04644.1 GBHO01017278 JAG26326.1 AEMK02000045 DQIR01206637 HDB62114.1 GDHC01016374 JAQ02255.1 ACTA01146508 ACTA01154507 UFQS01001903 UFQT01001903 SSX12672.1 SSX32115.1 GL193633 EFB13521.1 GBHO01032202 JAG11402.1 BC129706 AAI29707.1 AJVK01009891 KA646496 AFP61125.1 AAEX03009566 AJ875416 CAI44937.1 KV467236 OCT57062.1 GGFJ01007921 MBW57062.1
PCG78348.1 MF706190 ATJ44617.1 KQ459590 KPI97267.1 MF687646 ATJ44572.1 AK401487 BAM18109.1 KU755516 ARD71226.1 KJ579215 AIN34691.1 ODYU01004769 SOQ45001.1 KZ149984 PZC75714.1 AGBW02008350 OWR53605.1 GDQN01006492 JAT84562.1 KB632168 ERL89413.1 BT127049 AEE62011.1 GEBQ01008324 JAT31653.1 NNAY01004605 OXU17666.1 NIVC01003373 PAA51713.1 GEZM01029597 JAV86058.1 NEVH01013243 PNF29443.1 NIVC01001242 PAA70398.1 NIVC01001300 PAA69680.1 GECZ01016308 GECZ01005157 GECZ01001578 GECZ01000089 JAS53461.1 JAS64612.1 JAS68191.1 JAS69680.1 KK852567 KDR21202.1 GFDF01005633 JAV08451.1 GFDF01005632 JAV08452.1 GFDF01005638 JAV08446.1 GFDF01005637 JAV08447.1 KQ971319 EFA11379.1 AFYH01052445 AFYH01052446 UFQS01002445 UFQT01002445 SSX14055.1 SSX33474.1 GG666610 EEN49613.1 GFDL01007254 JAV27791.1 DS231824 EDS27414.1 AAMC01027184 AAMC01027185 BC076997 CR848336 AAH76997.1 CAJ83332.1 BC129713 AAI29714.1 CM004468 OCT94947.1 CH477470 EAT40438.1 DS469603 EDO39662.1 CH478592 EAT32741.1 AY455918 AAS16918.1 GAMD01001680 JAA99910.1 AMQN01003821 AMQN01003822 KB292015 ELU18250.1 GL762091 EFZ22264.1 AHAT01009296 GFFW01002564 JAV42224.1 AAAB01008852 EAA07412.4 APCN01009249 AGTP01008444 AGTP01008445 KF143809 AGV76999.1 GDRN01075798 JAI62990.1 LJIG01009943 KRT82069.1 GEDC01030531 JAS06767.1 GDHC01020622 GDHC01005136 JAP98006.1 JAQ13493.1 JXUM01046406 GAPW01002907 KQ561475 JAC10691.1 KXJ78478.1 GDHC01013294 JAQ05335.1 JXUM01060399 KQ562100 KXJ76693.1 GAMC01004539 JAC02017.1 CH940651 EDW65461.2 AEYP01007624 AEYP01007625 JP016046 AES04644.1 GBHO01017278 JAG26326.1 AEMK02000045 DQIR01206637 HDB62114.1 GDHC01016374 JAQ02255.1 ACTA01146508 ACTA01154507 UFQS01001903 UFQT01001903 SSX12672.1 SSX32115.1 GL193633 EFB13521.1 GBHO01032202 JAG11402.1 BC129706 AAI29707.1 AJVK01009891 KA646496 AFP61125.1 AAEX03009566 AJ875416 CAI44937.1 KV467236 OCT57062.1 GGFJ01007921 MBW57062.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000030742
UP000215335
+ More
UP000002358 UP000215902 UP000235965 UP000095280 UP000027135 UP000007266 UP000008672 UP000001554 UP000075884 UP000002320 UP000008143 UP000186698 UP000008820 UP000261560 UP000001593 UP000014760 UP000075885 UP000076407 UP000018468 UP000007062 UP000192223 UP000189704 UP000245300 UP000075840 UP000265120 UP000005215 UP000261520 UP000075903 UP000076408 UP000069940 UP000249989 UP000008792 UP000000715 UP000008227 UP000008912 UP000248482 UP000261680 UP000291021 UP000286642 UP000007635 UP000092462 UP000264840 UP000265080 UP000002254 UP000245340 UP000095301 UP000261580 UP000007646
UP000002358 UP000215902 UP000235965 UP000095280 UP000027135 UP000007266 UP000008672 UP000001554 UP000075884 UP000002320 UP000008143 UP000186698 UP000008820 UP000261560 UP000001593 UP000014760 UP000075885 UP000076407 UP000018468 UP000007062 UP000192223 UP000189704 UP000245300 UP000075840 UP000265120 UP000005215 UP000261520 UP000075903 UP000076408 UP000069940 UP000249989 UP000008792 UP000000715 UP000008227 UP000008912 UP000248482 UP000261680 UP000291021 UP000286642 UP000007635 UP000092462 UP000264840 UP000265080 UP000002254 UP000245340 UP000095301 UP000261580 UP000007646
PRIDE
Pfam
PF02089 Palm_thioest
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JQM9
W5VH72
D2SNX5
A0A2A4K346
A0A291P140
A0A194PVB8
+ More
A0A291P109 I4DJL9 A0A1V0M8B3 A0A088M9W1 A0A2H1VVX5 A0A2W1BQ24 A0A212FIN8 A0A1E1WC38 U4UAS9 J3JUY4 A0A1B6M6X5 A0A232EH28 K7IXU0 A0A267DQY9 A0A1Y1MLU8 A0A2J7QLI3 A0A1I8HQQ4 A0A1I8HWS3 A0A1B6H0L4 A0A067RBT8 A0A1L8DPX5 A0A1L8DPR4 A0A1L8DPR5 A0A1L8DPZ2 D6W6C0 H3B5N4 A0A336MSF6 C3ZCW0 A0A1Q3FJJ0 A0A182NDS4 B0W1X8 F6USM5 Q6DEU7 A2VDB5 A0A1L8HFT8 A0A1S4G3D1 Q170Q3 A0A1S4FHM6 A0A3B3BHH4 A7S9F2 Q16ET0 Q5WPS7 T1DPS5 R7VM55 A0A182PDH7 A0A1Y9J0D5 E9IAX5 W5LXU3 A0A250YFA7 Q7QDD6 A0A1W4XJH9 A0A1U7V115 S4RY76 A0A182IEP5 A0A3P8VM80 I3M5E3 T2FF50 A0A0P4WEX6 A0A0T6B4G0 A0A3B4BKW8 A0A182UVS8 A0A182YEM3 A0A1B6BZS0 A0A146M132 A0A023EP03 A0A146LCT3 A0A182GFT6 W8C624 B4M1A7 M3YS55 G9KIB9 A0A0A9Y3V5 A0A287AYH9 A0A146L3N2 G1LCS6 A0A336LF62 A0A2Y9IUU8 A0A384DT43 A0A3Q7XYH0 D2HXM1 G3NDR8 A0A0A9WSG7 A1L2U1 A0A1B0D0F5 A0A3Q2UST9 T1PCN1 A0A3P8U2Q8 Q5JZR0 A0A2U3X096 A0A1I8NAF5 A0A310UCB0 A0A3Q4HLX5 G3TPC5 A0A2M4BVN7
A0A291P109 I4DJL9 A0A1V0M8B3 A0A088M9W1 A0A2H1VVX5 A0A2W1BQ24 A0A212FIN8 A0A1E1WC38 U4UAS9 J3JUY4 A0A1B6M6X5 A0A232EH28 K7IXU0 A0A267DQY9 A0A1Y1MLU8 A0A2J7QLI3 A0A1I8HQQ4 A0A1I8HWS3 A0A1B6H0L4 A0A067RBT8 A0A1L8DPX5 A0A1L8DPR4 A0A1L8DPR5 A0A1L8DPZ2 D6W6C0 H3B5N4 A0A336MSF6 C3ZCW0 A0A1Q3FJJ0 A0A182NDS4 B0W1X8 F6USM5 Q6DEU7 A2VDB5 A0A1L8HFT8 A0A1S4G3D1 Q170Q3 A0A1S4FHM6 A0A3B3BHH4 A7S9F2 Q16ET0 Q5WPS7 T1DPS5 R7VM55 A0A182PDH7 A0A1Y9J0D5 E9IAX5 W5LXU3 A0A250YFA7 Q7QDD6 A0A1W4XJH9 A0A1U7V115 S4RY76 A0A182IEP5 A0A3P8VM80 I3M5E3 T2FF50 A0A0P4WEX6 A0A0T6B4G0 A0A3B4BKW8 A0A182UVS8 A0A182YEM3 A0A1B6BZS0 A0A146M132 A0A023EP03 A0A146LCT3 A0A182GFT6 W8C624 B4M1A7 M3YS55 G9KIB9 A0A0A9Y3V5 A0A287AYH9 A0A146L3N2 G1LCS6 A0A336LF62 A0A2Y9IUU8 A0A384DT43 A0A3Q7XYH0 D2HXM1 G3NDR8 A0A0A9WSG7 A1L2U1 A0A1B0D0F5 A0A3Q2UST9 T1PCN1 A0A3P8U2Q8 Q5JZR0 A0A2U3X096 A0A1I8NAF5 A0A310UCB0 A0A3Q4HLX5 G3TPC5 A0A2M4BVN7
PDB
3GRO
E-value=4.08848e-94,
Score=878
Ontologies
PATHWAY
GO
GO:0008474
GO:0002084
GO:0016740
GO:0006898
GO:0005764
GO:0051181
GO:0015031
GO:0008306
GO:0016042
GO:0030424
GO:0030425
GO:0007601
GO:0008021
GO:0048549
GO:0005615
GO:0007625
GO:0030308
GO:0032429
GO:0043025
GO:0051186
GO:0007420
GO:0007042
GO:0031579
GO:0005634
GO:0044257
GO:0048260
GO:0045121
GO:0016290
GO:0008344
GO:0005794
GO:0007269
GO:0043524
GO:0006907
GO:0005576
GO:0016021
GO:1900244
GO:0006897
GO:0048665
GO:1902667
GO:0008340
GO:0007399
GO:0043066
GO:0005829
GO:0098599
PANTHER
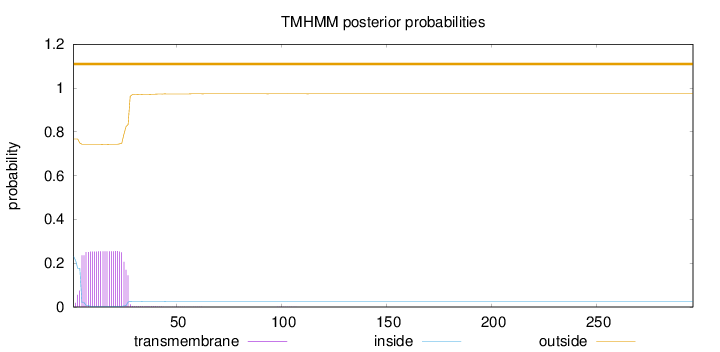
Topology
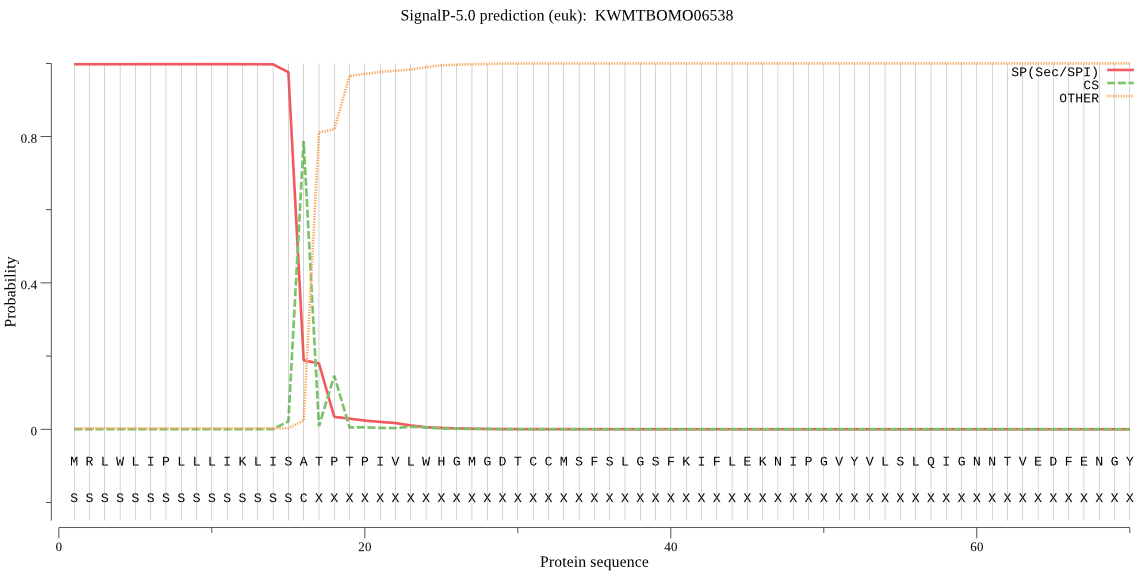
SignalP
Position: 1 - 16,
Likelihood: 0.997397
Length:
296
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.76778999999999
Exp number, first 60 AAs:
5.76452
Total prob of N-in:
0.23226
outside
1 - 296
Population Genetic Test Statistics
Pi
285.240286
Theta
182.997764
Tajima's D
1.453858
CLR
0.420504
CSRT
0.773661316934153
Interpretation
Uncertain
