Gene
KWMTBOMO06535 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011646
Annotation
PREDICTED:_beta-N-acetylglucosaminidase_1_isoform_X1_[Bombyx_mori]
Full name
Beta-hexosaminidase
Location in the cell
Cytoplasmic Reliability : 1.073
Sequence
CDS
ATGGCGCTTCATGGGCTGTTGGCGTTGATGACGACCATGCATTTGGCAGGTGTACTATGCAATGTAGAAAGTACATTTGACGAAATCACCCCTCAAATTTACGAACCATCATGGATGTATAAGTGCGTGCCAGATGAAGGATGTCAGCGTAGTGAACATCCTAGGCCGACATTGTCCGACAATTCAACGTCAGCTTTCTTCGATTCACTTGACGTGTGCCGTATAGTCTGCGGAAGGTTCGGCGGAATCTGGCCTAAGCCGGTTACAGCAGCACTGAGCTCGCAGACTGTGAAGATCCACCCTAATTATTTGAGATATGATCTGCTCAACGTGCCTGCAGAAACTAGGGAGCTATTAGTGGAAATGACGCAAGTGATAAGCAATAACTTGTTGGCTGAGTGCGGAGGACACGTCACTGAGGTGGTGGACACTCAGGTCGTTGTCATCATCGTGGTGAAGACCGCCATCACTTCGTTGAATTGGAACACTGACGAACAATACATGCTGGACGTACAAACTAGAGGAGGAGAAGTCTCTGTCCATATAGAAGCAGAAACCATTTACGGAGCTCGTCACGGCTTAGAGACCTTCTCTCAATTAATATCATCAGACAAACGTGACTTTTCTGATGTGGAACATTGCGGTTTGGTCCTCGTGTCCGGTGCAAAGATTCGTGACAGACCGATCTACAAACATAGAGGCTTAGTTCTAGATACTTCTAGACATTTCATACCGATGGTCGACATTAAAAGAACTATCGATGGTATGGCGACAACAAAAATGAATGTCTTTCATTGGCATGCCACCGATTCTCACAGTTTTCCTTTGGAAGCGAGTCGGGTGCCTCAGTTTACAAGATATGGCGCCTACTCGGGCAGCGAAATGTACACGACGGAAGAAATACGAGAGTTGATACATTACGCGAAGGTACGCGGCATCCGAGTCGTCATTGAAATAGACGCGCCGGCGCATTCAGGAAACGGATGGCAATGGGGAAGGGAATACGGCTTAGGTGATTTAGCGGTATGCGTTAACGCTTACCCCTGGAGACATCTTTGCATCGAACCTCCTTGTGGTCAACTGAATCCAGCGAACCCTAATATGTACCGGGTGCTGAGGAACTTGTACCAAGATGTTGCGGATCTGTTGAATTCACCGCCATTGCTACACATGGGAGGAGACGAGGTTTACTTCGGGTGCTGGAACTCGAGCCAAGAAATTATCTCGTACATGAAGGACCAAGGATACGATACGACGGAAGAAGGTTTCATGAAGTTGTGGGGTGAATTCCATAACAAAGCCCTTCAAATTTGGGACGAAGAGATTTCCGCGAAAGGACTGGACCCACAGCCCGTGATGTTATGGTCTTCGCAACTGACACAGGCTCAGAGGATCTCACAACACTTGGACAAAGAAAGATACATCATTGAAGTTTGGGAGCCTTTGAACAGCCCATTATTGACTCAACTCTTGAGGCTCGGCTATAGAACTGTTTCAGTACCGAAGGACATTTGGTATTTGGATCATGGCTTCTGGGGCAGAACGGTATATTCCAATTGGAGAAGGATGTATGCTCACACTCTACCACGGGACGAAGGCGTGTTAGGCGGTGAAGTGGCCATGTGGACGGAGTATTGCGACGCTCAGGCCCTAGACACCCGAGTATGGCCGAGAGCGGCTGCAGTGGCGGAGAGGCTGTGGTCAGACCCGACGTCCACAGTGTACAGCGCAGAGCCGAGGCTGCAGAGGCTCCGGACGAGACTGATAGCTCGGGGCCTGAGGCCCGACGCCATGTCCCCGGCGTGGTGCTCCCAGCACGACAGCAAATGTCTTTGA
Protein
MALHGLLALMTTMHLAGVLCNVESTFDEITPQIYEPSWMYKCVPDEGCQRSEHPRPTLSDNSTSAFFDSLDVCRIVCGRFGGIWPKPVTAALSSQTVKIHPNYLRYDLLNVPAETRELLVEMTQVISNNLLAECGGHVTEVVDTQVVVIIVVKTAITSLNWNTDEQYMLDVQTRGGEVSVHIEAETIYGARHGLETFSQLISSDKRDFSDVEHCGLVLVSGAKIRDRPIYKHRGLVLDTSRHFIPMVDIKRTIDGMATTKMNVFHWHATDSHSFPLEASRVPQFTRYGAYSGSEMYTTEEIRELIHYAKVRGIRVVIEIDAPAHSGNGWQWGREYGLGDLAVCVNAYPWRHLCIEPPCGQLNPANPNMYRVLRNLYQDVADLLNSPPLLHMGGDEVYFGCWNSSQEIISYMKDQGYDTTEEGFMKLWGEFHNKALQIWDEEISAKGLDPQPVMLWSSQLTQAQRISQHLDKERYIIEVWEPLNSPLLTQLLRLGYRTVSVPKDIWYLDHGFWGRTVYSNWRRMYAHTLPRDEGVLGGEVAMWTEYCDAQALDTRVWPRAAAVAERLWSDPTSTVYSAEPRLQRLRTRLIARGLRPDAMSPAWCSQHDSKCL
Summary
Catalytic Activity
Hydrolysis of terminal non-reducing N-acetyl-D-hexosamine residues in N-acetyl-beta-D-hexosaminides.
Similarity
Belongs to the glycosyl hydrolase 20 family.
Feature
chain Beta-hexosaminidase
Uniprot
A4PHN6
H9JQ40
A8IDA5
A0A212EXD1
A0A2W1BL06
A0A2A4K369
+ More
A0A194PW86 A0A2H1WAB3 A0A194QQN5 A0A0L7LUX1 A0A1Y1L8L6 A0A1Y1LDC0 A0A2J7QLP0 A0A2J7QLN5 A0A2J7QLQ5 A0A1B0CCX8 A0A1W4XJ86 A0A1W4XUF0 A0A1W4XU97 A0A1W4XTD2 A0A0C5C9K5 A0A1B6ED71 A0A182FQH0 A0A2P0P9N4 A0A2R7W0Q5 A0A1A9ZA56 A0A1B0FD25 A0A1J1IPT6 A0A1B0C609 A0A1A9XAV1 A0A2P8YWA3 A0A1Q3FIK6 A0A1I8N7F0 A0A1A9V5G4 A0A224XGY4 T1PAH1 A0A0M4EIH8 A0A0L0CIG2 A0A182GFT8 B4M1A8 A0A3B0KPR4 Q170Q1 E0VGG8 A0A1W4XJ90 A0A0Q9WD14 A0A067RNC2 B0W1X6 A5YVX4 Q29GS4 A0A1A9X5F5 B3MZG5 B4L3B1 B4JLT4 B4MJ44 T1HH13 A0A0A1WXP1 W8BGT9 A0A0K8UL98 Q7QKY7 A0A2H8TX73 Q9W3C4 B3NUL2 A0A1W4UQS0 A0A182MUW2 B4Q014 B4IK00 J9KAK0 A0A232F251 A0A336MD78 A0A1B0DGZ0 A0A1I8P8U7 K7IXT8 B4GV37 A0A182VPZ9 B5DN57 B4GUV2 A0A0A9Y3Z2 A0A0K8SIH6 A0A2K8JRH0 A0A0A9Y9Z5 A0A146KMB2 R4G8V6 A0A1Q1NPB5 A0A2S2NQI9 A0A1B6KVW3 A0A2H8U0P4 A0A182IRJ1 A0A182NN29 A0A3B0JXI5 A0A182G6N2 J9JVG2 A0A1A9WED7 A0A1A9ZAI9 A0A182QJ60 A0A1B6JM15 A0A1B0AU47 A0A1B0FRG6 A0A226F3N7 U4USN2
A0A194PW86 A0A2H1WAB3 A0A194QQN5 A0A0L7LUX1 A0A1Y1L8L6 A0A1Y1LDC0 A0A2J7QLP0 A0A2J7QLN5 A0A2J7QLQ5 A0A1B0CCX8 A0A1W4XJ86 A0A1W4XUF0 A0A1W4XU97 A0A1W4XTD2 A0A0C5C9K5 A0A1B6ED71 A0A182FQH0 A0A2P0P9N4 A0A2R7W0Q5 A0A1A9ZA56 A0A1B0FD25 A0A1J1IPT6 A0A1B0C609 A0A1A9XAV1 A0A2P8YWA3 A0A1Q3FIK6 A0A1I8N7F0 A0A1A9V5G4 A0A224XGY4 T1PAH1 A0A0M4EIH8 A0A0L0CIG2 A0A182GFT8 B4M1A8 A0A3B0KPR4 Q170Q1 E0VGG8 A0A1W4XJ90 A0A0Q9WD14 A0A067RNC2 B0W1X6 A5YVX4 Q29GS4 A0A1A9X5F5 B3MZG5 B4L3B1 B4JLT4 B4MJ44 T1HH13 A0A0A1WXP1 W8BGT9 A0A0K8UL98 Q7QKY7 A0A2H8TX73 Q9W3C4 B3NUL2 A0A1W4UQS0 A0A182MUW2 B4Q014 B4IK00 J9KAK0 A0A232F251 A0A336MD78 A0A1B0DGZ0 A0A1I8P8U7 K7IXT8 B4GV37 A0A182VPZ9 B5DN57 B4GUV2 A0A0A9Y3Z2 A0A0K8SIH6 A0A2K8JRH0 A0A0A9Y9Z5 A0A146KMB2 R4G8V6 A0A1Q1NPB5 A0A2S2NQI9 A0A1B6KVW3 A0A2H8U0P4 A0A182IRJ1 A0A182NN29 A0A3B0JXI5 A0A182G6N2 J9JVG2 A0A1A9WED7 A0A1A9ZAI9 A0A182QJ60 A0A1B6JM15 A0A1B0AU47 A0A1B0FRG6 A0A226F3N7 U4USN2
EC Number
3.2.1.52
Pubmed
17617724
19121390
22118469
28756777
26354079
26227816
+ More
28004739 25224926 29403074 25315136 26108605 26483478 17994087 17510324 20566863 24845553 18342252 18362917 19820115 15632085 25830018 24495485 9087549 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28648823 20075255 25401762 26823975 28130397 23537049
28004739 25224926 29403074 25315136 26108605 26483478 17994087 17510324 20566863 24845553 18342252 18362917 19820115 15632085 25830018 24495485 9087549 12364791 14747013 17210077 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28648823 20075255 25401762 26823975 28130397 23537049
EMBL
AB286957
BAF52531.1
BABH01012162
BABH01012163
EU158179
ABV79901.1
+ More
AGBW02011808 OWR46107.1 KZ149984 PZC75719.1 NWSH01000214 PCG78344.1 KQ459590 KPI97263.1 ODYU01007035 SOQ49424.1 KQ461190 KPJ07280.1 JTDY01000039 KOB79240.1 GEZM01062226 JAV69944.1 GEZM01062225 GEZM01062224 JAV69945.1 NEVH01013243 PNF29505.1 PNF29504.1 PNF29506.1 AJWK01007208 KM217123 AJO25048.1 GEDC01001410 JAS35888.1 KU739385 ARA90562.1 KK854169 PTY12510.1 CCAG010013058 CVRI01000055 CRL01556.1 JXJN01024611 JXJN01026390 PYGN01000323 PSN48529.1 GFDL01007625 JAV27420.1 GFTR01007348 JAW09078.1 KA645732 KA649460 AFP60361.1 CP012528 ALC48704.1 JRES01000348 KNC32035.1 JXUM01060403 KQ562100 KXJ76695.1 CH940651 EDW65462.1 OUUW01000015 SPP88609.1 CH477470 EAT40440.1 DS235142 EEB12474.1 KRF82242.1 KK852567 KDR21204.1 DS231824 EDS27412.1 EF592537 KQ971319 ABQ95983.1 EFA11380.2 CH379064 EAL32035.1 CH902635 EDV33766.1 CH933810 EDW07039.1 CH916371 EDV91695.1 CH963738 EDW72133.1 ACPB03022325 ACPB03022326 GBXI01012067 GBXI01010881 GBXI01005670 GBXI01005108 JAD02225.1 JAD03411.1 JAD08622.1 JAD09184.1 GAMC01010352 JAB96203.1 GDHF01024835 GDHF01014117 GDHF01013427 GDHF01003974 JAI27479.1 JAI38197.1 JAI38887.1 JAI48340.1 AAAB01008181 EAA03285.4 GFXV01007088 MBW18893.1 AE014298 AY118361 AAF46406.1 AAM48390.1 CH954180 EDV46335.1 AXCM01000572 CM000162 EDX02220.2 CH480850 EDW51356.1 ABLF02023225 ABLF02023226 NNAY01001235 OXU24662.1 UFQS01000959 UFQT01000959 SSX07952.1 SSX28186.1 AJVK01060576 CH479192 EDW26574.1 EDY72864.1 EDW26489.1 GBHO01017248 JAG26356.1 GBRD01012892 JAG52934.1 MF683372 ATU82513.1 GBHO01017249 JAG26355.1 GDHC01021744 JAP96884.1 GAHY01000307 JAA77203.1 KX459597 AQM58348.1 GGMR01006397 MBY19016.1 GEBQ01024365 GEBQ01023473 JAT15612.1 JAT16504.1 GFXV01007173 MBW18978.1 OUUW01000003 SPP78479.1 JXUM01046409 JXUM01046410 KQ561475 KXJ78479.1 ABLF02020377 AXCN02002216 GECU01007405 JAT00302.1 JXJN01003514 CCAG010017058 LNIX01000001 OXA63801.1 KB632432 ERL95528.1
AGBW02011808 OWR46107.1 KZ149984 PZC75719.1 NWSH01000214 PCG78344.1 KQ459590 KPI97263.1 ODYU01007035 SOQ49424.1 KQ461190 KPJ07280.1 JTDY01000039 KOB79240.1 GEZM01062226 JAV69944.1 GEZM01062225 GEZM01062224 JAV69945.1 NEVH01013243 PNF29505.1 PNF29504.1 PNF29506.1 AJWK01007208 KM217123 AJO25048.1 GEDC01001410 JAS35888.1 KU739385 ARA90562.1 KK854169 PTY12510.1 CCAG010013058 CVRI01000055 CRL01556.1 JXJN01024611 JXJN01026390 PYGN01000323 PSN48529.1 GFDL01007625 JAV27420.1 GFTR01007348 JAW09078.1 KA645732 KA649460 AFP60361.1 CP012528 ALC48704.1 JRES01000348 KNC32035.1 JXUM01060403 KQ562100 KXJ76695.1 CH940651 EDW65462.1 OUUW01000015 SPP88609.1 CH477470 EAT40440.1 DS235142 EEB12474.1 KRF82242.1 KK852567 KDR21204.1 DS231824 EDS27412.1 EF592537 KQ971319 ABQ95983.1 EFA11380.2 CH379064 EAL32035.1 CH902635 EDV33766.1 CH933810 EDW07039.1 CH916371 EDV91695.1 CH963738 EDW72133.1 ACPB03022325 ACPB03022326 GBXI01012067 GBXI01010881 GBXI01005670 GBXI01005108 JAD02225.1 JAD03411.1 JAD08622.1 JAD09184.1 GAMC01010352 JAB96203.1 GDHF01024835 GDHF01014117 GDHF01013427 GDHF01003974 JAI27479.1 JAI38197.1 JAI38887.1 JAI48340.1 AAAB01008181 EAA03285.4 GFXV01007088 MBW18893.1 AE014298 AY118361 AAF46406.1 AAM48390.1 CH954180 EDV46335.1 AXCM01000572 CM000162 EDX02220.2 CH480850 EDW51356.1 ABLF02023225 ABLF02023226 NNAY01001235 OXU24662.1 UFQS01000959 UFQT01000959 SSX07952.1 SSX28186.1 AJVK01060576 CH479192 EDW26574.1 EDY72864.1 EDW26489.1 GBHO01017248 JAG26356.1 GBRD01012892 JAG52934.1 MF683372 ATU82513.1 GBHO01017249 JAG26355.1 GDHC01021744 JAP96884.1 GAHY01000307 JAA77203.1 KX459597 AQM58348.1 GGMR01006397 MBY19016.1 GEBQ01024365 GEBQ01023473 JAT15612.1 JAT16504.1 GFXV01007173 MBW18978.1 OUUW01000003 SPP78479.1 JXUM01046409 JXUM01046410 KQ561475 KXJ78479.1 ABLF02020377 AXCN02002216 GECU01007405 JAT00302.1 JXJN01003514 CCAG010017058 LNIX01000001 OXA63801.1 KB632432 ERL95528.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000037510
+ More
UP000235965 UP000092461 UP000192223 UP000069272 UP000092445 UP000092444 UP000183832 UP000092460 UP000092443 UP000245037 UP000095301 UP000078200 UP000092553 UP000037069 UP000069940 UP000249989 UP000008792 UP000268350 UP000008820 UP000009046 UP000027135 UP000002320 UP000007266 UP000001819 UP000091820 UP000007801 UP000009192 UP000001070 UP000007798 UP000015103 UP000007062 UP000000803 UP000008711 UP000192221 UP000075883 UP000002282 UP000001292 UP000007819 UP000215335 UP000092462 UP000095300 UP000002358 UP000008744 UP000075920 UP000075880 UP000075884 UP000075886 UP000198287 UP000030742
UP000235965 UP000092461 UP000192223 UP000069272 UP000092445 UP000092444 UP000183832 UP000092460 UP000092443 UP000245037 UP000095301 UP000078200 UP000092553 UP000037069 UP000069940 UP000249989 UP000008792 UP000268350 UP000008820 UP000009046 UP000027135 UP000002320 UP000007266 UP000001819 UP000091820 UP000007801 UP000009192 UP000001070 UP000007798 UP000015103 UP000007062 UP000000803 UP000008711 UP000192221 UP000075883 UP000002282 UP000001292 UP000007819 UP000215335 UP000092462 UP000095300 UP000002358 UP000008744 UP000075920 UP000075880 UP000075884 UP000075886 UP000198287 UP000030742
Interpro
Gene 3D
ProteinModelPortal
A4PHN6
H9JQ40
A8IDA5
A0A212EXD1
A0A2W1BL06
A0A2A4K369
+ More
A0A194PW86 A0A2H1WAB3 A0A194QQN5 A0A0L7LUX1 A0A1Y1L8L6 A0A1Y1LDC0 A0A2J7QLP0 A0A2J7QLN5 A0A2J7QLQ5 A0A1B0CCX8 A0A1W4XJ86 A0A1W4XUF0 A0A1W4XU97 A0A1W4XTD2 A0A0C5C9K5 A0A1B6ED71 A0A182FQH0 A0A2P0P9N4 A0A2R7W0Q5 A0A1A9ZA56 A0A1B0FD25 A0A1J1IPT6 A0A1B0C609 A0A1A9XAV1 A0A2P8YWA3 A0A1Q3FIK6 A0A1I8N7F0 A0A1A9V5G4 A0A224XGY4 T1PAH1 A0A0M4EIH8 A0A0L0CIG2 A0A182GFT8 B4M1A8 A0A3B0KPR4 Q170Q1 E0VGG8 A0A1W4XJ90 A0A0Q9WD14 A0A067RNC2 B0W1X6 A5YVX4 Q29GS4 A0A1A9X5F5 B3MZG5 B4L3B1 B4JLT4 B4MJ44 T1HH13 A0A0A1WXP1 W8BGT9 A0A0K8UL98 Q7QKY7 A0A2H8TX73 Q9W3C4 B3NUL2 A0A1W4UQS0 A0A182MUW2 B4Q014 B4IK00 J9KAK0 A0A232F251 A0A336MD78 A0A1B0DGZ0 A0A1I8P8U7 K7IXT8 B4GV37 A0A182VPZ9 B5DN57 B4GUV2 A0A0A9Y3Z2 A0A0K8SIH6 A0A2K8JRH0 A0A0A9Y9Z5 A0A146KMB2 R4G8V6 A0A1Q1NPB5 A0A2S2NQI9 A0A1B6KVW3 A0A2H8U0P4 A0A182IRJ1 A0A182NN29 A0A3B0JXI5 A0A182G6N2 J9JVG2 A0A1A9WED7 A0A1A9ZAI9 A0A182QJ60 A0A1B6JM15 A0A1B0AU47 A0A1B0FRG6 A0A226F3N7 U4USN2
A0A194PW86 A0A2H1WAB3 A0A194QQN5 A0A0L7LUX1 A0A1Y1L8L6 A0A1Y1LDC0 A0A2J7QLP0 A0A2J7QLN5 A0A2J7QLQ5 A0A1B0CCX8 A0A1W4XJ86 A0A1W4XUF0 A0A1W4XU97 A0A1W4XTD2 A0A0C5C9K5 A0A1B6ED71 A0A182FQH0 A0A2P0P9N4 A0A2R7W0Q5 A0A1A9ZA56 A0A1B0FD25 A0A1J1IPT6 A0A1B0C609 A0A1A9XAV1 A0A2P8YWA3 A0A1Q3FIK6 A0A1I8N7F0 A0A1A9V5G4 A0A224XGY4 T1PAH1 A0A0M4EIH8 A0A0L0CIG2 A0A182GFT8 B4M1A8 A0A3B0KPR4 Q170Q1 E0VGG8 A0A1W4XJ90 A0A0Q9WD14 A0A067RNC2 B0W1X6 A5YVX4 Q29GS4 A0A1A9X5F5 B3MZG5 B4L3B1 B4JLT4 B4MJ44 T1HH13 A0A0A1WXP1 W8BGT9 A0A0K8UL98 Q7QKY7 A0A2H8TX73 Q9W3C4 B3NUL2 A0A1W4UQS0 A0A182MUW2 B4Q014 B4IK00 J9KAK0 A0A232F251 A0A336MD78 A0A1B0DGZ0 A0A1I8P8U7 K7IXT8 B4GV37 A0A182VPZ9 B5DN57 B4GUV2 A0A0A9Y3Z2 A0A0K8SIH6 A0A2K8JRH0 A0A0A9Y9Z5 A0A146KMB2 R4G8V6 A0A1Q1NPB5 A0A2S2NQI9 A0A1B6KVW3 A0A2H8U0P4 A0A182IRJ1 A0A182NN29 A0A3B0JXI5 A0A182G6N2 J9JVG2 A0A1A9WED7 A0A1A9ZAI9 A0A182QJ60 A0A1B6JM15 A0A1B0AU47 A0A1B0FRG6 A0A226F3N7 U4USN2
PDB
3OZP
E-value=1.08525e-104,
Score=973
Ontologies
PATHWAY
00511
Other glycan degradation - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
00531 Glycosaminoglycan degradation - Bombyx mori (domestic silkworm)
00603 Glycosphingolipid biosynthesis - globo and isoglobo series - Bombyx mori (domestic silkworm)
00604 Glycosphingolipid biosynthesis - ganglio series - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04142 Lysosome - Bombyx mori (domestic silkworm)
00520 Amino sugar and nucleotide sugar metabolism - Bombyx mori (domestic silkworm)
00531 Glycosaminoglycan degradation - Bombyx mori (domestic silkworm)
00603 Glycosphingolipid biosynthesis - globo and isoglobo series - Bombyx mori (domestic silkworm)
00604 Glycosphingolipid biosynthesis - ganglio series - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
04142 Lysosome - Bombyx mori (domestic silkworm)
GO
Topology
Subcellular location
Nucleus
SignalP
Position: 1 - 20,
Likelihood: 0.978534
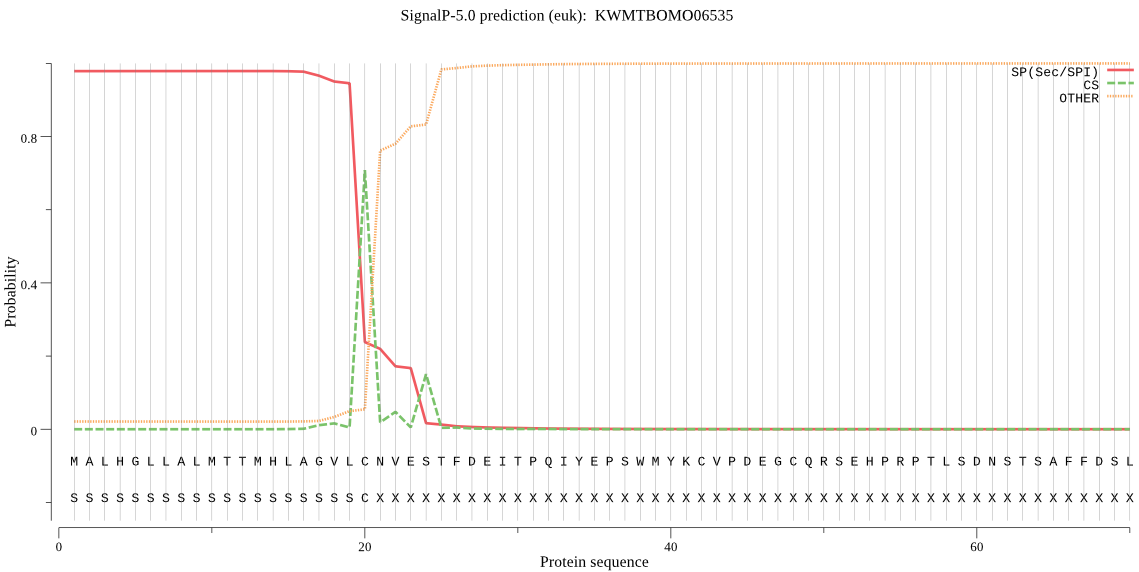
Length:
611
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0234
Exp number, first 60 AAs:
0.02269
Total prob of N-in:
0.00118
outside
1 - 611
Population Genetic Test Statistics
Pi
253.15854
Theta
201.607175
Tajima's D
1.10069
CLR
0.248388
CSRT
0.688015599220039
Interpretation
Uncertain
