Gene
KWMTBOMO06534
Annotation
PREDICTED:_protein_FAM195A_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.422
Sequence
CDS
ATGTACACCGTTTCCAAAGGACCCAGCAAGATTGTTGCAAAAACTAGAAGGGGCCTCTCCCAGAATCTAGAAAGGTTAGACAGTCGAAAAGATCATAACAGGAAGTCTTCAGATTCTGATACCGGAGAAATAATTAACATGCCTAGACCTATATTTCATTCCAATGGTAAAAAGACAGTGCATCAACGAATACAACATCATGTGATTACTCCACAGCACGAAGAAATAATTAGATTCATAAGTGAAACTTGGACACAATCAACATATGCAGACAGTGAACCCTCAACACCAACTAGTACGACTGATTCTGGCAGCTCATCCCCGGATCCACCCACCAATCTATACTACCAAGACGAAGCCCCAAGCCCCACACTAAGGGACTTCAAACCCTTCGACTTAGAAACGTGGTGGGGCAAACGACTTTTCCAGAACATAACAAACTCCCTTTGA
Protein
MYTVSKGPSKIVAKTRRGLSQNLERLDSRKDHNRKSSDSDTGEIINMPRPIFHSNGKKTVHQRIQHHVITPQHEEIIRFISETWTQSTYADSEPSTPTSTTDSGSSSPDPPTNLYYQDEAPSPTLRDFKPFDLETWWGKRLFQNITNSL
Summary
Uniprot
A0A2H1VRE9
A0A2W1BSI7
A0A2A4K3J9
A0A1E1W605
A0A194QUP9
A0A194PVB4
+ More
A0A212EX80 A0A1B6CSM9 A0A195FH17 A0A151XHE8 F4WSP0 A0A195DJ26 A0A195C0H1 A0A0L7QNC9 A0A087ZZE2 A0A2A3EIT2 A0A310SCH2 A0A151I0T3 A0A0M9A7C4 A0A026WWZ1 A0A1L8DCZ5 E2B6W3 A0A067R3Z2 E9IVA7 A0A232FM97 A0A0C9QKL8 A0A1A9UM11 D3TM86 A0A1A9ZDN2 A0A1I8Q7D1 A0A1I8MXM1 A0A1B6FPP2 A0A0L0C8M3 A0A1B6MEF0 A0A1A9W4D3 A0A1B6ID87 E2AFD7 A0A224Y0A7 A0A0V0GB52 A0A023F8V7 A0A069DP94 A0A2P8ZMC7 B4IU00 B4PCF9 A0A0P4VN53 R4G4D2 B3NI24 A0A1W4VP00 Q9VUP0 B4HI81 B4QL20 A0A1A9Y1K8 A0A1B0BPU9 Q2M1B8 B4GUC1 B3M711 A0A3B0L051 B4NM41 A0A0M4EKI8 T1DFA2 A0A182GKA3 Q17NK4 A0A1Q3FEA1 A0A2M3ZIU8 B4IZ09 A0A1Q3FEF5 A0A182WEY5 A0A1Y1M923 B4L150 W5JTS0 Q7Q0D1 A0A0A9YRV3 A0A182WZS8 B4LH70 A0A2M4C1D6 B0WFN5 A0A2M4AL03 A0A182RD01 A0A1W4XUA5 A0A0P5TCK6 E9H1M8 A0A182KA82 A0A023EIU3 D7EHQ5 U5EYY1 V5IAT5 A0A0P4WCP1 A0A1J1HIJ5 A0A087TTY5 A0A034W484 A0A0K8U9U7 B4G591 A0A0A1XLM0 B5DY42 A0A0K8TKN8 A0A3S3SF93 B4NFX3
A0A212EX80 A0A1B6CSM9 A0A195FH17 A0A151XHE8 F4WSP0 A0A195DJ26 A0A195C0H1 A0A0L7QNC9 A0A087ZZE2 A0A2A3EIT2 A0A310SCH2 A0A151I0T3 A0A0M9A7C4 A0A026WWZ1 A0A1L8DCZ5 E2B6W3 A0A067R3Z2 E9IVA7 A0A232FM97 A0A0C9QKL8 A0A1A9UM11 D3TM86 A0A1A9ZDN2 A0A1I8Q7D1 A0A1I8MXM1 A0A1B6FPP2 A0A0L0C8M3 A0A1B6MEF0 A0A1A9W4D3 A0A1B6ID87 E2AFD7 A0A224Y0A7 A0A0V0GB52 A0A023F8V7 A0A069DP94 A0A2P8ZMC7 B4IU00 B4PCF9 A0A0P4VN53 R4G4D2 B3NI24 A0A1W4VP00 Q9VUP0 B4HI81 B4QL20 A0A1A9Y1K8 A0A1B0BPU9 Q2M1B8 B4GUC1 B3M711 A0A3B0L051 B4NM41 A0A0M4EKI8 T1DFA2 A0A182GKA3 Q17NK4 A0A1Q3FEA1 A0A2M3ZIU8 B4IZ09 A0A1Q3FEF5 A0A182WEY5 A0A1Y1M923 B4L150 W5JTS0 Q7Q0D1 A0A0A9YRV3 A0A182WZS8 B4LH70 A0A2M4C1D6 B0WFN5 A0A2M4AL03 A0A182RD01 A0A1W4XUA5 A0A0P5TCK6 E9H1M8 A0A182KA82 A0A023EIU3 D7EHQ5 U5EYY1 V5IAT5 A0A0P4WCP1 A0A1J1HIJ5 A0A087TTY5 A0A034W484 A0A0K8U9U7 B4G591 A0A0A1XLM0 B5DY42 A0A0K8TKN8 A0A3S3SF93 B4NFX3
Pubmed
28756777
26354079
22118469
21719571
24508170
20798317
+ More
24845553 21282665 28648823 20353571 25315136 26108605 25474469 26334808 29403074 17994087 17550304 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 24330624 26483478 17510324 28004739 20920257 23761445 12364791 14747013 17210077 25401762 26823975 21292972 24945155 18362917 19820115 25348373 25830018 26369729
24845553 21282665 28648823 20353571 25315136 26108605 25474469 26334808 29403074 17994087 17550304 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 15632085 24330624 26483478 17510324 28004739 20920257 23761445 12364791 14747013 17210077 25401762 26823975 21292972 24945155 18362917 19820115 25348373 25830018 26369729
EMBL
ODYU01003982
SOQ43399.1
KZ149984
PZC75720.1
NWSH01000214
PCG78343.1
+ More
GDQN01008648 JAT82406.1 KQ461190 KPJ07281.1 KQ459590 KPI97262.1 AGBW02011808 OWR46106.1 GEDC01020897 GEDC01018294 JAS16401.1 JAS19004.1 KQ981560 KYN39980.1 KQ982130 KYQ59826.1 GL888326 EGI62783.1 KQ980800 KYN12822.1 KQ978379 KYM94399.1 KQ414855 KOC60147.1 KZ288230 PBC31598.1 KQ779356 OAD51973.1 KQ976599 KYM79502.1 KQ435732 KOX77359.1 KK107102 EZA59629.1 GFDF01009758 JAV04326.1 GL446031 EFN88568.1 KK852999 KDR12662.1 GL766194 EFZ15470.1 NNAY01000041 OXU31629.1 GBYB01004099 JAG73866.1 EZ422538 ADD18814.1 GECZ01017592 JAS52177.1 JRES01000760 KNC28582.1 GEBQ01005718 JAT34259.1 GECU01022827 JAS84879.1 GL439108 EFN67830.1 GFTR01002423 JAW14003.1 GECL01001156 JAP04968.1 GBBI01000841 JAC17871.1 GBGD01003383 JAC85506.1 PYGN01000017 PSN57643.1 CH891752 EDW99863.1 CM000159 EDW94869.1 GDKW01000551 JAI56044.1 ACPB03028992 GAHY01000871 JAA76639.1 CH954178 EDV52110.1 AE014296 AY084095 AAF49635.1 AAL89833.1 AGB94569.1 CH480815 EDW41577.1 CM000363 CM002912 EDX10545.1 KMY99744.1 JXJN01018237 CH379069 EAL30656.1 CH479191 EDW26204.1 CH902618 EDV40876.1 OUUW01000027 SPP89758.1 CH964278 EDW85409.1 CP012525 ALC44701.1 GALA01000707 JAA94145.1 JXUM01069652 JXUM01069653 JXUM01069654 JXUM01069655 KQ562562 KXJ75619.1 CH477198 EAT48311.1 GFDL01009158 JAV25887.1 GGFM01007746 MBW28497.1 CH916366 EDV97717.1 GFDL01009153 JAV25892.1 GEZM01042484 JAV79827.1 CH933809 EDW18207.1 ADMH02000120 ETN67757.1 AAAB01008986 EAA00527.4 GBHO01011344 GBRD01009795 GDHC01013976 JAG32260.1 JAG56029.1 JAQ04653.1 CH940647 EDW70583.1 GGFJ01009972 MBW59113.1 DS231920 DS233029 EDS26379.1 EDS27592.1 GGFK01008144 MBW41465.1 GDIQ01219573 GDIP01128012 LRGB01000024 JAK32152.1 JAL75702.1 KZS21553.1 GL732583 EFX74472.1 GAPW01004723 JAC08875.1 DS497665 EFA12137.1 GANO01000040 JAB59831.1 GALX01000625 JAB67841.1 GDRN01099674 GDRN01099673 JAI58729.1 CVRI01000006 CRK87872.1 KK116723 KFM68574.1 GAKP01009840 JAC49112.1 GDHF01028872 JAI23442.1 CH479179 EDW24757.1 GBXI01002043 JAD12249.1 CM000070 EDY67963.1 GDAI01002905 JAI14698.1 NCKU01001032 RWS13302.1 CH964251 EDW83190.1
GDQN01008648 JAT82406.1 KQ461190 KPJ07281.1 KQ459590 KPI97262.1 AGBW02011808 OWR46106.1 GEDC01020897 GEDC01018294 JAS16401.1 JAS19004.1 KQ981560 KYN39980.1 KQ982130 KYQ59826.1 GL888326 EGI62783.1 KQ980800 KYN12822.1 KQ978379 KYM94399.1 KQ414855 KOC60147.1 KZ288230 PBC31598.1 KQ779356 OAD51973.1 KQ976599 KYM79502.1 KQ435732 KOX77359.1 KK107102 EZA59629.1 GFDF01009758 JAV04326.1 GL446031 EFN88568.1 KK852999 KDR12662.1 GL766194 EFZ15470.1 NNAY01000041 OXU31629.1 GBYB01004099 JAG73866.1 EZ422538 ADD18814.1 GECZ01017592 JAS52177.1 JRES01000760 KNC28582.1 GEBQ01005718 JAT34259.1 GECU01022827 JAS84879.1 GL439108 EFN67830.1 GFTR01002423 JAW14003.1 GECL01001156 JAP04968.1 GBBI01000841 JAC17871.1 GBGD01003383 JAC85506.1 PYGN01000017 PSN57643.1 CH891752 EDW99863.1 CM000159 EDW94869.1 GDKW01000551 JAI56044.1 ACPB03028992 GAHY01000871 JAA76639.1 CH954178 EDV52110.1 AE014296 AY084095 AAF49635.1 AAL89833.1 AGB94569.1 CH480815 EDW41577.1 CM000363 CM002912 EDX10545.1 KMY99744.1 JXJN01018237 CH379069 EAL30656.1 CH479191 EDW26204.1 CH902618 EDV40876.1 OUUW01000027 SPP89758.1 CH964278 EDW85409.1 CP012525 ALC44701.1 GALA01000707 JAA94145.1 JXUM01069652 JXUM01069653 JXUM01069654 JXUM01069655 KQ562562 KXJ75619.1 CH477198 EAT48311.1 GFDL01009158 JAV25887.1 GGFM01007746 MBW28497.1 CH916366 EDV97717.1 GFDL01009153 JAV25892.1 GEZM01042484 JAV79827.1 CH933809 EDW18207.1 ADMH02000120 ETN67757.1 AAAB01008986 EAA00527.4 GBHO01011344 GBRD01009795 GDHC01013976 JAG32260.1 JAG56029.1 JAQ04653.1 CH940647 EDW70583.1 GGFJ01009972 MBW59113.1 DS231920 DS233029 EDS26379.1 EDS27592.1 GGFK01008144 MBW41465.1 GDIQ01219573 GDIP01128012 LRGB01000024 JAK32152.1 JAL75702.1 KZS21553.1 GL732583 EFX74472.1 GAPW01004723 JAC08875.1 DS497665 EFA12137.1 GANO01000040 JAB59831.1 GALX01000625 JAB67841.1 GDRN01099674 GDRN01099673 JAI58729.1 CVRI01000006 CRK87872.1 KK116723 KFM68574.1 GAKP01009840 JAC49112.1 GDHF01028872 JAI23442.1 CH479179 EDW24757.1 GBXI01002043 JAD12249.1 CM000070 EDY67963.1 GDAI01002905 JAI14698.1 NCKU01001032 RWS13302.1 CH964251 EDW83190.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000078541
UP000075809
+ More
UP000007755 UP000078492 UP000078542 UP000053825 UP000005203 UP000242457 UP000078540 UP000053105 UP000053097 UP000008237 UP000027135 UP000215335 UP000078200 UP000092445 UP000095300 UP000095301 UP000037069 UP000091820 UP000000311 UP000245037 UP000002282 UP000015103 UP000008711 UP000192221 UP000000803 UP000001292 UP000000304 UP000092443 UP000092460 UP000001819 UP000008744 UP000007801 UP000268350 UP000007798 UP000092553 UP000069940 UP000249989 UP000008820 UP000001070 UP000075920 UP000009192 UP000000673 UP000007062 UP000076407 UP000008792 UP000002320 UP000075900 UP000192223 UP000076858 UP000000305 UP000075881 UP000007266 UP000183832 UP000054359 UP000285301
UP000007755 UP000078492 UP000078542 UP000053825 UP000005203 UP000242457 UP000078540 UP000053105 UP000053097 UP000008237 UP000027135 UP000215335 UP000078200 UP000092445 UP000095300 UP000095301 UP000037069 UP000091820 UP000000311 UP000245037 UP000002282 UP000015103 UP000008711 UP000192221 UP000000803 UP000001292 UP000000304 UP000092443 UP000092460 UP000001819 UP000008744 UP000007801 UP000268350 UP000007798 UP000092553 UP000069940 UP000249989 UP000008820 UP000001070 UP000075920 UP000009192 UP000000673 UP000007062 UP000076407 UP000008792 UP000002320 UP000075900 UP000192223 UP000076858 UP000000305 UP000075881 UP000007266 UP000183832 UP000054359 UP000285301
PRIDE
Pfam
PF14799 FAM195
Interpro
IPR029428
MCRIP
ProteinModelPortal
A0A2H1VRE9
A0A2W1BSI7
A0A2A4K3J9
A0A1E1W605
A0A194QUP9
A0A194PVB4
+ More
A0A212EX80 A0A1B6CSM9 A0A195FH17 A0A151XHE8 F4WSP0 A0A195DJ26 A0A195C0H1 A0A0L7QNC9 A0A087ZZE2 A0A2A3EIT2 A0A310SCH2 A0A151I0T3 A0A0M9A7C4 A0A026WWZ1 A0A1L8DCZ5 E2B6W3 A0A067R3Z2 E9IVA7 A0A232FM97 A0A0C9QKL8 A0A1A9UM11 D3TM86 A0A1A9ZDN2 A0A1I8Q7D1 A0A1I8MXM1 A0A1B6FPP2 A0A0L0C8M3 A0A1B6MEF0 A0A1A9W4D3 A0A1B6ID87 E2AFD7 A0A224Y0A7 A0A0V0GB52 A0A023F8V7 A0A069DP94 A0A2P8ZMC7 B4IU00 B4PCF9 A0A0P4VN53 R4G4D2 B3NI24 A0A1W4VP00 Q9VUP0 B4HI81 B4QL20 A0A1A9Y1K8 A0A1B0BPU9 Q2M1B8 B4GUC1 B3M711 A0A3B0L051 B4NM41 A0A0M4EKI8 T1DFA2 A0A182GKA3 Q17NK4 A0A1Q3FEA1 A0A2M3ZIU8 B4IZ09 A0A1Q3FEF5 A0A182WEY5 A0A1Y1M923 B4L150 W5JTS0 Q7Q0D1 A0A0A9YRV3 A0A182WZS8 B4LH70 A0A2M4C1D6 B0WFN5 A0A2M4AL03 A0A182RD01 A0A1W4XUA5 A0A0P5TCK6 E9H1M8 A0A182KA82 A0A023EIU3 D7EHQ5 U5EYY1 V5IAT5 A0A0P4WCP1 A0A1J1HIJ5 A0A087TTY5 A0A034W484 A0A0K8U9U7 B4G591 A0A0A1XLM0 B5DY42 A0A0K8TKN8 A0A3S3SF93 B4NFX3
A0A212EX80 A0A1B6CSM9 A0A195FH17 A0A151XHE8 F4WSP0 A0A195DJ26 A0A195C0H1 A0A0L7QNC9 A0A087ZZE2 A0A2A3EIT2 A0A310SCH2 A0A151I0T3 A0A0M9A7C4 A0A026WWZ1 A0A1L8DCZ5 E2B6W3 A0A067R3Z2 E9IVA7 A0A232FM97 A0A0C9QKL8 A0A1A9UM11 D3TM86 A0A1A9ZDN2 A0A1I8Q7D1 A0A1I8MXM1 A0A1B6FPP2 A0A0L0C8M3 A0A1B6MEF0 A0A1A9W4D3 A0A1B6ID87 E2AFD7 A0A224Y0A7 A0A0V0GB52 A0A023F8V7 A0A069DP94 A0A2P8ZMC7 B4IU00 B4PCF9 A0A0P4VN53 R4G4D2 B3NI24 A0A1W4VP00 Q9VUP0 B4HI81 B4QL20 A0A1A9Y1K8 A0A1B0BPU9 Q2M1B8 B4GUC1 B3M711 A0A3B0L051 B4NM41 A0A0M4EKI8 T1DFA2 A0A182GKA3 Q17NK4 A0A1Q3FEA1 A0A2M3ZIU8 B4IZ09 A0A1Q3FEF5 A0A182WEY5 A0A1Y1M923 B4L150 W5JTS0 Q7Q0D1 A0A0A9YRV3 A0A182WZS8 B4LH70 A0A2M4C1D6 B0WFN5 A0A2M4AL03 A0A182RD01 A0A1W4XUA5 A0A0P5TCK6 E9H1M8 A0A182KA82 A0A023EIU3 D7EHQ5 U5EYY1 V5IAT5 A0A0P4WCP1 A0A1J1HIJ5 A0A087TTY5 A0A034W484 A0A0K8U9U7 B4G591 A0A0A1XLM0 B5DY42 A0A0K8TKN8 A0A3S3SF93 B4NFX3
Ontologies
GO
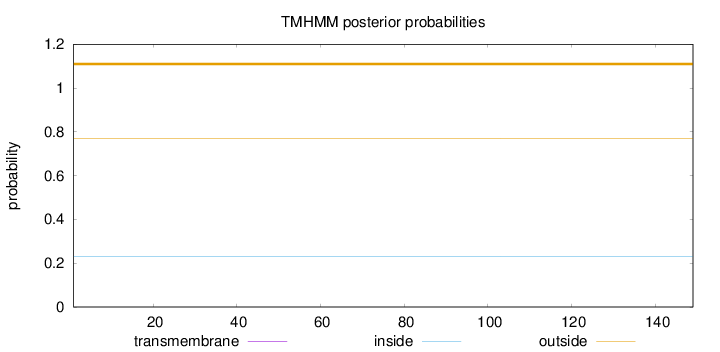
Topology
Length:
149
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.22922
outside
1 - 149
Population Genetic Test Statistics
Pi
248.57901
Theta
203.971206
Tajima's D
0.66935
CLR
0.375528
CSRT
0.566271686415679
Interpretation
Uncertain
