Gene
KWMTBOMO06533
Pre Gene Modal
BGIBMGA011645
Annotation
PREDICTED:_mpv17-like_protein_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 3.294
Sequence
CDS
ATGGCGTCAAAACTACTGGCTTGGTCGCGCCAATCACTACAACGTCGTCCGGTTCTAACCAACACCGTAGTCTACGCAACGTTTTATACTGCCGCTGAATTGTCGCAGCAAACCTTCAATAGAATTTACTCGCCGGAAAAGCCAGATTTAGATGTCGCGGCTGCCGCCCGAATCGTCTCAGTTGGTAGTACTGTTTACGCACCCACATTGTACTTTTGGTACAAATTTTTGGACAAGAAGTTTGCCGGCAACGCGATCAAAGCCGTAGCGACAAAAGTGGCTTCCGATCAGTTCATTATGACGCCTATATTATTGGCCACGTTTTATACTTTGTTGGGGATTTTAGAACGTAGAGAAGATGTTTTCGAAGAGCTAAAGCAGAAATACTGGAAGACGTTTATAGCCAATCAAGCATTTTGGATTCCAGCGCAAACGATTAACTTTTACTTCATGCCTTCACATTTGAGAGTTGTTTACGTAGCCTCAACATCTTTCGTGTGGATCAATATCCTTTGCTTTATTAAGAGACAAAAAAACATGAAAAAACTTGAAGAGATGCCATTATAA
Protein
MASKLLAWSRQSLQRRPVLTNTVVYATFYTAAELSQQTFNRIYSPEKPDLDVAAAARIVSVGSTVYAPTLYFWYKFLDKKFAGNAIKAVATKVASDQFIMTPILLATFYTLLGILERREDVFEELKQKYWKTFIANQAFWIPAQTINFYFMPSHLRVVYVASTSFVWINILCFIKRQKNMKKLEEMPL
Summary
Similarity
Belongs to the peroxisomal membrane protein PXMP2/4 family.
Uniprot
H9JQ39
A0A194PWX1
A0A194QPK4
A0A1E1W5Z0
A0A212EXD8
A0A1E1WK29
+ More
A0A3S2NGU6 A0A2A4K244 A0A2H1VRC6 A0A0L7LEW2 A0A1D2N4K9 A0A0A1WHC6 A0A0L0C813 A0A139WDG6 Q17NK0 A0A0A9XRX9 A0A034WKG4 A0A226E3J5 A0A182QIW8 A0A182HQC1 A0A1Q3G545 W8B480 B4LH64 A0A182WZS6 Q7Q0C9 A0A087UP74 A0A0K8WL19 A0A1B6HHJ7 A0A182KSB7 A0A182USE1 A0A182PKA8 A0A182RFH2 A0A182M194 A0A0Q9XBZ2 A0A182Y1I4 A0A1B0GQF1 A0A1B6FHG2 B4L144 A0A1J1I600 A0A1B6L1H5 A0A0M4EFG2 R4G3B0 A0A0P8YIM2 A0A1S3DAQ6 A0A182FB38 J3JVJ8 A0A182WEY7 A0A3B0K6L7 A0A1B0FUX6 A0A023F7S6 A0A336MAQ7 A0A336MUH5 A0A1I8PKH1 B5DQH9 B4GUC7 A0A1W4VN52 B4QL15 A0A1L8D9B9 W5JW51 A0A1B6ECR2 A0A131XYT0 B4ITZ4 A0A1I8MRS6 A0A1A9WFE6 A0A2H8TPV1 A0A2J7Q8Q5 A0A139WD90 A0A3R7QC64 A0A1A9VMK9 A8JNT1 C4WRZ8 B3NI18 A0A1B0G7G6 A0A2S2PZD8 A0A1A9Z358 A0A0P4WL76 A0A2L2YVE1 A0A1S3DC47 A0A293MS31 A0A1E1XJ41 A0A0K8VYB7 A0A0P4W7L1 L7LS54 A0A1V9XCM3 V5GQQ9 A0A3R7PS75 L7MGK1 G3MPX4 A0A224Z3F1 A0A131YC70 A0A131XJF7 L7M4J8 A0A0L7QSE6 A0A1S3KFE9 R7UPB4 A0A182H3E4 E2AVA6 A0A2L2YMJ1
A0A3S2NGU6 A0A2A4K244 A0A2H1VRC6 A0A0L7LEW2 A0A1D2N4K9 A0A0A1WHC6 A0A0L0C813 A0A139WDG6 Q17NK0 A0A0A9XRX9 A0A034WKG4 A0A226E3J5 A0A182QIW8 A0A182HQC1 A0A1Q3G545 W8B480 B4LH64 A0A182WZS6 Q7Q0C9 A0A087UP74 A0A0K8WL19 A0A1B6HHJ7 A0A182KSB7 A0A182USE1 A0A182PKA8 A0A182RFH2 A0A182M194 A0A0Q9XBZ2 A0A182Y1I4 A0A1B0GQF1 A0A1B6FHG2 B4L144 A0A1J1I600 A0A1B6L1H5 A0A0M4EFG2 R4G3B0 A0A0P8YIM2 A0A1S3DAQ6 A0A182FB38 J3JVJ8 A0A182WEY7 A0A3B0K6L7 A0A1B0FUX6 A0A023F7S6 A0A336MAQ7 A0A336MUH5 A0A1I8PKH1 B5DQH9 B4GUC7 A0A1W4VN52 B4QL15 A0A1L8D9B9 W5JW51 A0A1B6ECR2 A0A131XYT0 B4ITZ4 A0A1I8MRS6 A0A1A9WFE6 A0A2H8TPV1 A0A2J7Q8Q5 A0A139WD90 A0A3R7QC64 A0A1A9VMK9 A8JNT1 C4WRZ8 B3NI18 A0A1B0G7G6 A0A2S2PZD8 A0A1A9Z358 A0A0P4WL76 A0A2L2YVE1 A0A1S3DC47 A0A293MS31 A0A1E1XJ41 A0A0K8VYB7 A0A0P4W7L1 L7LS54 A0A1V9XCM3 V5GQQ9 A0A3R7PS75 L7MGK1 G3MPX4 A0A224Z3F1 A0A131YC70 A0A131XJF7 L7M4J8 A0A0L7QSE6 A0A1S3KFE9 R7UPB4 A0A182H3E4 E2AVA6 A0A2L2YMJ1
Pubmed
19121390
26354079
22118469
26227816
27289101
25830018
+ More
26108605 18362917 19820115 17510324 25401762 26823975 25348373 24495485 17994087 12364791 14747013 17210077 20966253 25244985 22516182 25474469 15632085 22936249 20920257 23761445 29652888 17550304 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26561354 29209593 25576852 28327890 25765539 22216098 28797301 26830274 28049606 23254933 26483478 20798317
26108605 18362917 19820115 17510324 25401762 26823975 25348373 24495485 17994087 12364791 14747013 17210077 20966253 25244985 22516182 25474469 15632085 22936249 20920257 23761445 29652888 17550304 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26561354 29209593 25576852 28327890 25765539 22216098 28797301 26830274 28049606 23254933 26483478 20798317
EMBL
BABH01012163
BABH01012164
KQ459590
KPI97259.1
KQ461190
KPJ07284.1
+ More
GDQN01008695 JAT82359.1 AGBW02011808 OWR46104.1 GDQN01003808 JAT87246.1 RSAL01000043 RVE50723.1 NWSH01000214 PCG78341.1 ODYU01003982 SOQ43401.1 JTDY01001353 KOB74098.1 LJIJ01000224 ODN00203.1 GBXI01016201 JAC98090.1 JRES01000760 KNC28583.1 KQ971361 KYB25895.1 CH477198 EAT48315.1 GBHO01023759 GBRD01005265 GBRD01005263 GDHC01000708 JAG19845.1 JAG60556.1 JAQ17921.1 GAKP01004327 JAC54625.1 LNIX01000007 OXA51537.1 AXCN02001024 APCN01003306 GFDL01000115 JAV34930.1 GAMC01014687 JAB91868.1 CH940647 EDW70577.1 AAAB01008986 EAA00178.4 KK120837 KFM79163.1 GDHF01000446 JAI51868.1 GECU01033533 GECU01031067 JAS74173.1 JAS76639.1 AXCM01005723 CH933809 KRG06064.1 AJVK01007171 AJVK01007172 GECZ01020131 GECZ01019122 JAS49638.1 JAS50647.1 EDW18201.1 CVRI01000042 CRK95751.1 GEBQ01030985 GEBQ01022593 JAT08992.1 JAT17384.1 CP012525 ALC44684.1 ACPB03014724 GAHY01001841 JAA75669.1 CH902618 KPU78826.1 BT127266 AEE62228.1 OUUW01000027 SPP89764.1 AJWK01006385 AJWK01006386 AJWK01006387 GBBI01001453 JAC17259.1 UFQS01000827 UFQT01000827 SSX07079.1 SSX27422.1 UFQS01002674 UFQT01002674 SSX14501.1 SSX33906.1 CH379069 EDY73370.1 CH479191 EDW26210.1 CM000363 CM002912 EDX10540.1 KMY99736.1 GFDF01011015 JAV03069.1 ADMH02000120 ETN67758.1 GEDC01027397 GEDC01001572 JAS09901.1 JAS35726.1 GEFM01004516 GEGO01000040 JAP71280.1 JAR95364.1 CM000159 CH891752 EDW94863.1 EDW99857.1 GFXV01004391 MBW16196.1 NEVH01016949 PNF24962.1 KYB25896.1 QCYY01001922 ROT74217.1 AE014296 ABW08543.1 AGB94568.1 ABLF02029752 AK339984 BAH70668.1 CH954178 EDV52104.1 CCAG010000131 GGMS01001570 MBY70773.1 GDRN01064184 JAI64883.1 IAAA01060902 LAA11997.1 GFWV01018598 MAA43326.1 GFAA01004104 JAT99330.1 GDHF01008485 JAI43829.1 GDRN01065858 JAI64629.1 GACK01010642 JAA54392.1 MNPL01014991 OQR71284.1 GANP01011713 JAB72755.1 QCYY01001788 ROT75302.1 GACK01002636 JAA62398.1 JO843925 AEO35542.1 GFPF01009947 MAA21093.1 GEDV01012060 JAP76497.1 GEFH01002331 JAP66250.1 GACK01006242 JAA58792.1 KQ414758 KOC61474.1 AMQN01007897 KB301692 ELU05241.1 JXUM01107321 JXUM01107322 JXUM01107323 JXUM01107324 JXUM01107325 KQ565131 KXJ71302.1 GL443059 EFN62660.1 IAAA01038074 LAA09299.1
GDQN01008695 JAT82359.1 AGBW02011808 OWR46104.1 GDQN01003808 JAT87246.1 RSAL01000043 RVE50723.1 NWSH01000214 PCG78341.1 ODYU01003982 SOQ43401.1 JTDY01001353 KOB74098.1 LJIJ01000224 ODN00203.1 GBXI01016201 JAC98090.1 JRES01000760 KNC28583.1 KQ971361 KYB25895.1 CH477198 EAT48315.1 GBHO01023759 GBRD01005265 GBRD01005263 GDHC01000708 JAG19845.1 JAG60556.1 JAQ17921.1 GAKP01004327 JAC54625.1 LNIX01000007 OXA51537.1 AXCN02001024 APCN01003306 GFDL01000115 JAV34930.1 GAMC01014687 JAB91868.1 CH940647 EDW70577.1 AAAB01008986 EAA00178.4 KK120837 KFM79163.1 GDHF01000446 JAI51868.1 GECU01033533 GECU01031067 JAS74173.1 JAS76639.1 AXCM01005723 CH933809 KRG06064.1 AJVK01007171 AJVK01007172 GECZ01020131 GECZ01019122 JAS49638.1 JAS50647.1 EDW18201.1 CVRI01000042 CRK95751.1 GEBQ01030985 GEBQ01022593 JAT08992.1 JAT17384.1 CP012525 ALC44684.1 ACPB03014724 GAHY01001841 JAA75669.1 CH902618 KPU78826.1 BT127266 AEE62228.1 OUUW01000027 SPP89764.1 AJWK01006385 AJWK01006386 AJWK01006387 GBBI01001453 JAC17259.1 UFQS01000827 UFQT01000827 SSX07079.1 SSX27422.1 UFQS01002674 UFQT01002674 SSX14501.1 SSX33906.1 CH379069 EDY73370.1 CH479191 EDW26210.1 CM000363 CM002912 EDX10540.1 KMY99736.1 GFDF01011015 JAV03069.1 ADMH02000120 ETN67758.1 GEDC01027397 GEDC01001572 JAS09901.1 JAS35726.1 GEFM01004516 GEGO01000040 JAP71280.1 JAR95364.1 CM000159 CH891752 EDW94863.1 EDW99857.1 GFXV01004391 MBW16196.1 NEVH01016949 PNF24962.1 KYB25896.1 QCYY01001922 ROT74217.1 AE014296 ABW08543.1 AGB94568.1 ABLF02029752 AK339984 BAH70668.1 CH954178 EDV52104.1 CCAG010000131 GGMS01001570 MBY70773.1 GDRN01064184 JAI64883.1 IAAA01060902 LAA11997.1 GFWV01018598 MAA43326.1 GFAA01004104 JAT99330.1 GDHF01008485 JAI43829.1 GDRN01065858 JAI64629.1 GACK01010642 JAA54392.1 MNPL01014991 OQR71284.1 GANP01011713 JAB72755.1 QCYY01001788 ROT75302.1 GACK01002636 JAA62398.1 JO843925 AEO35542.1 GFPF01009947 MAA21093.1 GEDV01012060 JAP76497.1 GEFH01002331 JAP66250.1 GACK01006242 JAA58792.1 KQ414758 KOC61474.1 AMQN01007897 KB301692 ELU05241.1 JXUM01107321 JXUM01107322 JXUM01107323 JXUM01107324 JXUM01107325 KQ565131 KXJ71302.1 GL443059 EFN62660.1 IAAA01038074 LAA09299.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000283053
UP000218220
+ More
UP000037510 UP000094527 UP000037069 UP000007266 UP000008820 UP000198287 UP000075886 UP000075840 UP000008792 UP000076407 UP000007062 UP000054359 UP000075882 UP000075903 UP000075885 UP000075900 UP000075883 UP000009192 UP000076408 UP000092462 UP000183832 UP000092553 UP000015103 UP000007801 UP000079169 UP000069272 UP000075920 UP000268350 UP000092461 UP000095300 UP000001819 UP000008744 UP000192221 UP000000304 UP000000673 UP000002282 UP000095301 UP000091820 UP000235965 UP000283509 UP000078200 UP000000803 UP000007819 UP000008711 UP000092444 UP000092445 UP000192247 UP000053825 UP000085678 UP000014760 UP000069940 UP000249989 UP000000311
UP000037510 UP000094527 UP000037069 UP000007266 UP000008820 UP000198287 UP000075886 UP000075840 UP000008792 UP000076407 UP000007062 UP000054359 UP000075882 UP000075903 UP000075885 UP000075900 UP000075883 UP000009192 UP000076408 UP000092462 UP000183832 UP000092553 UP000015103 UP000007801 UP000079169 UP000069272 UP000075920 UP000268350 UP000092461 UP000095300 UP000001819 UP000008744 UP000192221 UP000000304 UP000000673 UP000002282 UP000095301 UP000091820 UP000235965 UP000283509 UP000078200 UP000000803 UP000007819 UP000008711 UP000092444 UP000092445 UP000192247 UP000053825 UP000085678 UP000014760 UP000069940 UP000249989 UP000000311
Interpro
SUPFAM
SSF55729
SSF55729
ProteinModelPortal
H9JQ39
A0A194PWX1
A0A194QPK4
A0A1E1W5Z0
A0A212EXD8
A0A1E1WK29
+ More
A0A3S2NGU6 A0A2A4K244 A0A2H1VRC6 A0A0L7LEW2 A0A1D2N4K9 A0A0A1WHC6 A0A0L0C813 A0A139WDG6 Q17NK0 A0A0A9XRX9 A0A034WKG4 A0A226E3J5 A0A182QIW8 A0A182HQC1 A0A1Q3G545 W8B480 B4LH64 A0A182WZS6 Q7Q0C9 A0A087UP74 A0A0K8WL19 A0A1B6HHJ7 A0A182KSB7 A0A182USE1 A0A182PKA8 A0A182RFH2 A0A182M194 A0A0Q9XBZ2 A0A182Y1I4 A0A1B0GQF1 A0A1B6FHG2 B4L144 A0A1J1I600 A0A1B6L1H5 A0A0M4EFG2 R4G3B0 A0A0P8YIM2 A0A1S3DAQ6 A0A182FB38 J3JVJ8 A0A182WEY7 A0A3B0K6L7 A0A1B0FUX6 A0A023F7S6 A0A336MAQ7 A0A336MUH5 A0A1I8PKH1 B5DQH9 B4GUC7 A0A1W4VN52 B4QL15 A0A1L8D9B9 W5JW51 A0A1B6ECR2 A0A131XYT0 B4ITZ4 A0A1I8MRS6 A0A1A9WFE6 A0A2H8TPV1 A0A2J7Q8Q5 A0A139WD90 A0A3R7QC64 A0A1A9VMK9 A8JNT1 C4WRZ8 B3NI18 A0A1B0G7G6 A0A2S2PZD8 A0A1A9Z358 A0A0P4WL76 A0A2L2YVE1 A0A1S3DC47 A0A293MS31 A0A1E1XJ41 A0A0K8VYB7 A0A0P4W7L1 L7LS54 A0A1V9XCM3 V5GQQ9 A0A3R7PS75 L7MGK1 G3MPX4 A0A224Z3F1 A0A131YC70 A0A131XJF7 L7M4J8 A0A0L7QSE6 A0A1S3KFE9 R7UPB4 A0A182H3E4 E2AVA6 A0A2L2YMJ1
A0A3S2NGU6 A0A2A4K244 A0A2H1VRC6 A0A0L7LEW2 A0A1D2N4K9 A0A0A1WHC6 A0A0L0C813 A0A139WDG6 Q17NK0 A0A0A9XRX9 A0A034WKG4 A0A226E3J5 A0A182QIW8 A0A182HQC1 A0A1Q3G545 W8B480 B4LH64 A0A182WZS6 Q7Q0C9 A0A087UP74 A0A0K8WL19 A0A1B6HHJ7 A0A182KSB7 A0A182USE1 A0A182PKA8 A0A182RFH2 A0A182M194 A0A0Q9XBZ2 A0A182Y1I4 A0A1B0GQF1 A0A1B6FHG2 B4L144 A0A1J1I600 A0A1B6L1H5 A0A0M4EFG2 R4G3B0 A0A0P8YIM2 A0A1S3DAQ6 A0A182FB38 J3JVJ8 A0A182WEY7 A0A3B0K6L7 A0A1B0FUX6 A0A023F7S6 A0A336MAQ7 A0A336MUH5 A0A1I8PKH1 B5DQH9 B4GUC7 A0A1W4VN52 B4QL15 A0A1L8D9B9 W5JW51 A0A1B6ECR2 A0A131XYT0 B4ITZ4 A0A1I8MRS6 A0A1A9WFE6 A0A2H8TPV1 A0A2J7Q8Q5 A0A139WD90 A0A3R7QC64 A0A1A9VMK9 A8JNT1 C4WRZ8 B3NI18 A0A1B0G7G6 A0A2S2PZD8 A0A1A9Z358 A0A0P4WL76 A0A2L2YVE1 A0A1S3DC47 A0A293MS31 A0A1E1XJ41 A0A0K8VYB7 A0A0P4W7L1 L7LS54 A0A1V9XCM3 V5GQQ9 A0A3R7PS75 L7MGK1 G3MPX4 A0A224Z3F1 A0A131YC70 A0A131XJF7 L7M4J8 A0A0L7QSE6 A0A1S3KFE9 R7UPB4 A0A182H3E4 E2AVA6 A0A2L2YMJ1
Ontologies
GO
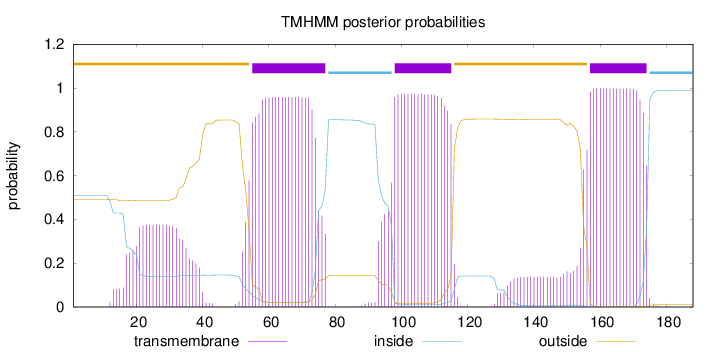
Topology
Length:
188
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
70.75552
Exp number, first 60 AAs:
14.37696
Total prob of N-in:
0.50866
POSSIBLE N-term signal
sequence
outside
1 - 54
TMhelix
55 - 77
inside
78 - 97
TMhelix
98 - 115
outside
116 - 156
TMhelix
157 - 174
inside
175 - 188
Population Genetic Test Statistics
Pi
222.772716
Theta
228.131731
Tajima's D
0.219903
CLR
0.52908
CSRT
0.434978251087446
Interpretation
Uncertain
