Gene
KWMTBOMO06530
Pre Gene Modal
BGIBMGA011644
Annotation
PREDICTED:_fas-binding_factor_1_homolog_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.183
Sequence
CDS
ATGCAAACTATCATAAATGCTCATCAAGAACGCATAAACCAGAGGATAAAGGCCCTCTTGGGAACTCAGGATACGATCGATGAAGAAGGACACGCTGATGGAGCTTTCGAAATGAATGTCGGTGACGGTAAAGAAACGCCTCAACGTAAGGAAAACAAACAGCTTTTGCAATTGATTCAGTCTCTGCAGGAGAATCACGATAAAGAAATTGATTTGATGGAAACGTCTTATAGACGCCAACTAGCGTTCTTGGAGCTATCGGTGCATCAATCTGAAGAGCGAATGAAGGAGGAAAGCGATAAACTGGTTAAGTTCTATTCGGAGAAGATCGGCTGGCTAGAGGAACATCACTTGCTATTCAAGAAGATGACTGAGGATAATTTGAGTGCGTTAACCGATAGGCATAAAGCTGAGAACGACATGCTGAGACACCAGCATTTGGACAACGTAAGGATCTTGCAGGAACACCACACCGCCCTTATGGAAAATATCAAGAATGCCGTGAAGCAGGAGCAGATCCTTATTAAGGATTCTGTTTCGTTCTCGTCTGATTTAAGTGATTTAGTGGCGGAAGTCAGGGAGACTAAAACTAACTGTCAATTACTTGTTAATAAAGTTGAAACTTTATCTGAAGCCAATCGCCATGATACCGAAAAGAGTTTGCAAACTAGAGAGACTCAGATTAATGATATGTTACAACACTTGAAAAGGGAGCGGGAAAATTTCGATATCGAAAAGAATGAAAGTCGAGATATTGTTAGAATGTTGGAAGCAAGACTAAAACAAATGACGGCGACGCTTGAAGAAGAAACGGCATTATTAAAACAGAAGAGAATGGAATTTGAATTTGAAAAAGCGACATTCAATAAGCAAACAGAATTCGCTAAGAACGTTCTGAAGAAACAAGACGACGAAATAAAAACGCTTAGGGAAGACATCCAAAAAGAATACCAGGAGAAGATCGCGAAGATAGACGAGGAAAAGGTTAAGACGTTAAAAGATAGTAGCGCGGTAGCAAAAGATAAGGCAGCCGTGCAGAGTTTAAAACAAGAACTTGAGAAAATGAAAGCGGAACTGCAAGCGCAACTTGAAGAACTGACTGAGGAGAGAACTAAGTTGAATGTTGAAAAGCAACAAGTGCACATGGAGGAACAGAGAACAGTGGCCAAGAGTCGTGATCTGGACTTGCTAGCCAAAACGGCGATTGAGAAGCAGTCCCAAGCCGACAAGAAATATTCTGAGGCGGAGTTCTTACAGAGAAAATACGAGGAGCGAATACGGAGGATACAGGAGCATGTGGTTTCGTTGAACGCCCGCGAGAAGCAGATCGCCAGAGAGAAGGTGGCTCTGTCCAGAGAAAGGCTGAACTTGCACAACGAGAGGAGGCAGATCGAGGGAAAGCAGTGTTCTCTGTGCAAATCCACCCAAAACTTACCTTTCAGTTTAGAAACTAACTATAACGTGCCAGATTCGTACCTTAATGTATCGGTTTCGAGGGATCCAACGAAGCCCAACGTGAGCTCGGCCATGAACGCCATCGAACAAGAGATGGCCCATTTGATGAGTGCTAATTTTAGTTTAAGACACACTCCCGGTGTTAGTGACGTAAGTGAAGTTAATAGGTTTGTTGGGGGCGAAGCTCTCGATGGTACAATACAAAATGCTGAATCTCAATTGGGATCTGGAGCTTTTAAGGATTACATGGACCCCAAGTTCATGATGTTACGTTTGGACGTTCAGAAAGTATTGAGTAATATTGATCAAAGTAAAAATGGCGATATTAAATACGGTCATTCCGATGAAGACTAA
Protein
MQTIINAHQERINQRIKALLGTQDTIDEEGHADGAFEMNVGDGKETPQRKENKQLLQLIQSLQENHDKEIDLMETSYRRQLAFLELSVHQSEERMKEESDKLVKFYSEKIGWLEEHHLLFKKMTEDNLSALTDRHKAENDMLRHQHLDNVRILQEHHTALMENIKNAVKQEQILIKDSVSFSSDLSDLVAEVRETKTNCQLLVNKVETLSEANRHDTEKSLQTRETQINDMLQHLKRERENFDIEKNESRDIVRMLEARLKQMTATLEEETALLKQKRMEFEFEKATFNKQTEFAKNVLKKQDDEIKTLREDIQKEYQEKIAKIDEEKVKTLKDSSAVAKDKAAVQSLKQELEKMKAELQAQLEELTEERTKLNVEKQQVHMEEQRTVAKSRDLDLLAKTAIEKQSQADKKYSEAEFLQRKYEERIRRIQEHVVSLNAREKQIAREKVALSRERLNLHNERRQIEGKQCSLCKSTQNLPFSLETNYNVPDSYLNVSVSRDPTKPNVSSAMNAIEQEMAHLMSANFSLRHTPGVSDVSEVNRFVGGEALDGTIQNAESQLGSGAFKDYMDPKFMMLRLDVQKVLSNIDQSKNGDIKYGHSDED
Summary
Uniprot
H9JQ38
A0A2A4J7L3
A0A2W1BQ32
A0A2H1X304
A0A3S2M3U9
A0A0L7LJU5
+ More
A0A194PV30 A0A194QP34 D6WHC4 W8BT00 W8AU28 A0A182W3U0 A0A0L0BSE6 A0A182RBB7 B0X7E5 A0A182QNF7 A0A182MTJ5 A0A1S4FEK1 Q174W7 A0A0T6BFK2 A0A0A1X6U4 A0A182H9U5 A0A182PI42 A0A182S6E4 A0A067R0L6 A0A182IYH0 A0A0K8W9S4 A0A2P8Z058 A0A1W4WMT0 A0A1W4WCS8 A0A1I8MQR2 A0A034V5B4 A0A182NDM3 A0A182YIQ1 A0A2J7R9W7 A0A182FM50 B4LCS7 A0A1S4H420 A0A182X856 A0A182LNB1 A0A182HXA7 Q7QDV8 A0A182VCI1 B4N734 A0A1I8NRW0 Q9VTN6 B4IZA5 A0A1B0F9X7 A0A2J7R9W8 A0A1A9YH02 K7IXZ6 A0A1B0BC64 A0A1A9X0E5 A0A232FGM2 A0A2K5C606 A0A0L7RJN3 A0A1A9ZTZ5 A0A2K5C624 A0A3B0J556 A0A026WE06 A0A2A3EIF0 A0A1A9UQK1 M3YQP2 A0A2C9LL54 H2NUR6 A0A2K5C5Z6 A0A2K5C5Y4 E2B4F4 A0A2J8Y134 A0A2J8Y150 A0A2J8Y130 A0A2C9LKY0
A0A194PV30 A0A194QP34 D6WHC4 W8BT00 W8AU28 A0A182W3U0 A0A0L0BSE6 A0A182RBB7 B0X7E5 A0A182QNF7 A0A182MTJ5 A0A1S4FEK1 Q174W7 A0A0T6BFK2 A0A0A1X6U4 A0A182H9U5 A0A182PI42 A0A182S6E4 A0A067R0L6 A0A182IYH0 A0A0K8W9S4 A0A2P8Z058 A0A1W4WMT0 A0A1W4WCS8 A0A1I8MQR2 A0A034V5B4 A0A182NDM3 A0A182YIQ1 A0A2J7R9W7 A0A182FM50 B4LCS7 A0A1S4H420 A0A182X856 A0A182LNB1 A0A182HXA7 Q7QDV8 A0A182VCI1 B4N734 A0A1I8NRW0 Q9VTN6 B4IZA5 A0A1B0F9X7 A0A2J7R9W8 A0A1A9YH02 K7IXZ6 A0A1B0BC64 A0A1A9X0E5 A0A232FGM2 A0A2K5C606 A0A0L7RJN3 A0A1A9ZTZ5 A0A2K5C624 A0A3B0J556 A0A026WE06 A0A2A3EIF0 A0A1A9UQK1 M3YQP2 A0A2C9LL54 H2NUR6 A0A2K5C5Z6 A0A2K5C5Y4 E2B4F4 A0A2J8Y134 A0A2J8Y150 A0A2J8Y130 A0A2C9LKY0
Pubmed
19121390
28756777
26227816
26354079
18362917
19820115
+ More
24495485 26108605 17510324 25830018 26483478 24845553 29403074 25315136 25348373 25244985 17994087 12364791 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20075255 28648823 24508170 30249741 15562597 20798317
24495485 26108605 17510324 25830018 26483478 24845553 29403074 25315136 25348373 25244985 17994087 12364791 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20075255 28648823 24508170 30249741 15562597 20798317
EMBL
BABH01012175
NWSH01002655
PCG67746.1
KZ149984
PZC75724.1
ODYU01013013
+ More
SOQ59618.1 RSAL01000043 RVE50725.1 JTDY01000862 KOB75634.1 KQ459590 KPI97256.1 KQ461190 KPJ07288.1 KQ971321 EFA00110.2 GAMC01014211 JAB92344.1 GAMC01014210 JAB92345.1 JRES01001542 KNC22154.1 DS232448 EDS41895.1 AXCN02001749 AXCM01001917 CH477405 EAT41597.1 LJIG01000856 KRT86084.1 GBXI01007248 JAD07044.1 JXUM01004840 KQ560182 KXJ83964.1 KK853032 KDR12259.1 GDHF01004403 JAI47911.1 PYGN01000259 PSN49874.1 GAKP01020466 JAC38486.1 NEVH01006580 PNF37629.1 CH940647 EDW70969.2 AAAB01008849 APCN01005168 EAA07126.4 CH964168 EDW80173.1 AE014296 AAF50010.1 CH916366 EDV96660.1 CCAG010008448 PNF37628.1 JXJN01011858 NNAY01000287 OXU29477.1 KQ414578 KOC71167.1 OUUW01000002 SPP76755.1 KK107260 QOIP01000001 EZA54148.1 RLU27207.1 KZ288253 PBC30979.1 AEYP01025312 AEYP01025313 AEYP01025314 ABGA01016335 ABGA01016336 ABGA01016337 ABGA01016338 ABGA01016339 ABGA01016340 ABGA01016341 ABGA01016342 GL445552 EFN89448.1 NDHI03003284 PNJ87978.1 PNJ87979.1 PNJ87980.1
SOQ59618.1 RSAL01000043 RVE50725.1 JTDY01000862 KOB75634.1 KQ459590 KPI97256.1 KQ461190 KPJ07288.1 KQ971321 EFA00110.2 GAMC01014211 JAB92344.1 GAMC01014210 JAB92345.1 JRES01001542 KNC22154.1 DS232448 EDS41895.1 AXCN02001749 AXCM01001917 CH477405 EAT41597.1 LJIG01000856 KRT86084.1 GBXI01007248 JAD07044.1 JXUM01004840 KQ560182 KXJ83964.1 KK853032 KDR12259.1 GDHF01004403 JAI47911.1 PYGN01000259 PSN49874.1 GAKP01020466 JAC38486.1 NEVH01006580 PNF37629.1 CH940647 EDW70969.2 AAAB01008849 APCN01005168 EAA07126.4 CH964168 EDW80173.1 AE014296 AAF50010.1 CH916366 EDV96660.1 CCAG010008448 PNF37628.1 JXJN01011858 NNAY01000287 OXU29477.1 KQ414578 KOC71167.1 OUUW01000002 SPP76755.1 KK107260 QOIP01000001 EZA54148.1 RLU27207.1 KZ288253 PBC30979.1 AEYP01025312 AEYP01025313 AEYP01025314 ABGA01016335 ABGA01016336 ABGA01016337 ABGA01016338 ABGA01016339 ABGA01016340 ABGA01016341 ABGA01016342 GL445552 EFN89448.1 NDHI03003284 PNJ87978.1 PNJ87979.1 PNJ87980.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000037510
UP000053268
UP000053240
+ More
UP000007266 UP000075920 UP000037069 UP000075900 UP000002320 UP000075886 UP000075883 UP000008820 UP000069940 UP000249989 UP000075885 UP000075901 UP000027135 UP000075880 UP000245037 UP000192223 UP000095301 UP000075884 UP000076408 UP000235965 UP000069272 UP000008792 UP000076407 UP000075882 UP000075840 UP000007062 UP000075903 UP000007798 UP000095300 UP000000803 UP000001070 UP000092444 UP000092443 UP000002358 UP000092460 UP000091820 UP000215335 UP000233020 UP000053825 UP000092445 UP000268350 UP000053097 UP000279307 UP000242457 UP000078200 UP000000715 UP000076420 UP000001595 UP000008237
UP000007266 UP000075920 UP000037069 UP000075900 UP000002320 UP000075886 UP000075883 UP000008820 UP000069940 UP000249989 UP000075885 UP000075901 UP000027135 UP000075880 UP000245037 UP000192223 UP000095301 UP000075884 UP000076408 UP000235965 UP000069272 UP000008792 UP000076407 UP000075882 UP000075840 UP000007062 UP000075903 UP000007798 UP000095300 UP000000803 UP000001070 UP000092444 UP000092443 UP000002358 UP000092460 UP000091820 UP000215335 UP000233020 UP000053825 UP000092445 UP000268350 UP000053097 UP000279307 UP000242457 UP000078200 UP000000715 UP000076420 UP000001595 UP000008237
Interpro
IPR033561
FBF1
ProteinModelPortal
H9JQ38
A0A2A4J7L3
A0A2W1BQ32
A0A2H1X304
A0A3S2M3U9
A0A0L7LJU5
+ More
A0A194PV30 A0A194QP34 D6WHC4 W8BT00 W8AU28 A0A182W3U0 A0A0L0BSE6 A0A182RBB7 B0X7E5 A0A182QNF7 A0A182MTJ5 A0A1S4FEK1 Q174W7 A0A0T6BFK2 A0A0A1X6U4 A0A182H9U5 A0A182PI42 A0A182S6E4 A0A067R0L6 A0A182IYH0 A0A0K8W9S4 A0A2P8Z058 A0A1W4WMT0 A0A1W4WCS8 A0A1I8MQR2 A0A034V5B4 A0A182NDM3 A0A182YIQ1 A0A2J7R9W7 A0A182FM50 B4LCS7 A0A1S4H420 A0A182X856 A0A182LNB1 A0A182HXA7 Q7QDV8 A0A182VCI1 B4N734 A0A1I8NRW0 Q9VTN6 B4IZA5 A0A1B0F9X7 A0A2J7R9W8 A0A1A9YH02 K7IXZ6 A0A1B0BC64 A0A1A9X0E5 A0A232FGM2 A0A2K5C606 A0A0L7RJN3 A0A1A9ZTZ5 A0A2K5C624 A0A3B0J556 A0A026WE06 A0A2A3EIF0 A0A1A9UQK1 M3YQP2 A0A2C9LL54 H2NUR6 A0A2K5C5Z6 A0A2K5C5Y4 E2B4F4 A0A2J8Y134 A0A2J8Y150 A0A2J8Y130 A0A2C9LKY0
A0A194PV30 A0A194QP34 D6WHC4 W8BT00 W8AU28 A0A182W3U0 A0A0L0BSE6 A0A182RBB7 B0X7E5 A0A182QNF7 A0A182MTJ5 A0A1S4FEK1 Q174W7 A0A0T6BFK2 A0A0A1X6U4 A0A182H9U5 A0A182PI42 A0A182S6E4 A0A067R0L6 A0A182IYH0 A0A0K8W9S4 A0A2P8Z058 A0A1W4WMT0 A0A1W4WCS8 A0A1I8MQR2 A0A034V5B4 A0A182NDM3 A0A182YIQ1 A0A2J7R9W7 A0A182FM50 B4LCS7 A0A1S4H420 A0A182X856 A0A182LNB1 A0A182HXA7 Q7QDV8 A0A182VCI1 B4N734 A0A1I8NRW0 Q9VTN6 B4IZA5 A0A1B0F9X7 A0A2J7R9W8 A0A1A9YH02 K7IXZ6 A0A1B0BC64 A0A1A9X0E5 A0A232FGM2 A0A2K5C606 A0A0L7RJN3 A0A1A9ZTZ5 A0A2K5C624 A0A3B0J556 A0A026WE06 A0A2A3EIF0 A0A1A9UQK1 M3YQP2 A0A2C9LL54 H2NUR6 A0A2K5C5Z6 A0A2K5C5Y4 E2B4F4 A0A2J8Y134 A0A2J8Y150 A0A2J8Y130 A0A2C9LKY0
Ontologies
GO
PANTHER
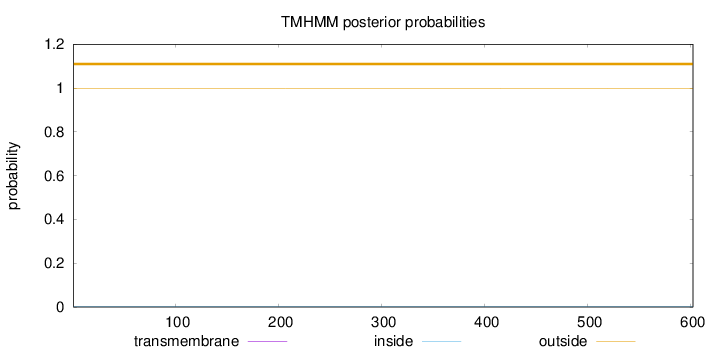
Topology
Length:
602
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00156
outside
1 - 602
Population Genetic Test Statistics
Pi
187.810438
Theta
162.597009
Tajima's D
0.376417
CLR
19.94899
CSRT
0.47667616619169
Interpretation
Uncertain
