Gene
KWMTBOMO06527
Pre Gene Modal
BGIBMGA014606
Annotation
PREDICTED:_uncharacterized_protein_LOC101741787_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.906
Sequence
CDS
ATGTTTAAAAGACTGTATGAAGAGACTGGAGGTAGCTATGAGAGCTTGGATGACGGATGTCAATTCTTGAATTTTAAAACTGACTTGTTTAGCTTAAAGGAAGTTTTCAATATGCCTGAAGAAAGAGTGAAAAATGAACCAGGACAAAAACCTTGGGGTAACTGTCATCCAGAAATACTATCAAAGGTCAGACAGTATTACAATGTACCAGATTTCTTACCAAGTGATGCTGAATTTTCTGGTACTGAAAATATTTTTTTCGGTTATGAAATTGGAGCAGTTATGCATTTGGATTACATATCAAGATTGATGTGGCAGGGTCAAGTAAGTGGTAACAAAATATGGTCTATTGCACCAGTACCAGAGTGTGATCATGTCTGTAATGGTTTGGAGTTTTATGTAGAAAAAGGTGACATAGTATTTTTGGACACTCGCATCTGGTATCATTCAACAAGGATTCCTGCAGGAGAATTTTCTTTAACTGTCCAATCAGAATATGGTTAA
Protein
MFKRLYEETGGSYESLDDGCQFLNFKTDLFSLKEVFNMPEERVKNEPGQKPWGNCHPEILSKVRQYYNVPDFLPSDAEFSGTENIFFGYEIGAVMHLDYISRLMWQGQVSGNKIWSIAPVPECDHVCNGLEFYVEKGDIVFLDTRIWYHSTRIPAGEFSLTVQSEYG
Summary
Uniprot
H9JYI6
A0A2A4J797
A0A2H1W2T7
A0A3S2LCM1
S4PSF5
A0A212FP60
+ More
A0A1W4W8H8 A0A194PWW6 A0A194QQP5 A0A232EPT5 A0A2J7Q1Z8 K7IM31 A0A310S8H2 E0VIT0 A0A067QZK3 A0A0N0U560 A0A151I5N8 A0A154PNP3 A0A0K8TCI3 A0A146M9Q1 A0A0A9XY89 E9IPK6 A0A195CK58 A0A195EVS3 A0A195DQJ3 A0A158NAJ9 A0A088ANZ2 A0A023F0J5 A0A0L7LJN2 D6WHD1 E2BPJ4 A0A0K8TQP7 E2ABK9 A0A069DRD9 A0A151X5E4 A0A026WWR9 A0A1B6HX94 A0A0L7QN51 A0A0V0GDD1 A0A1Y1K228 A0A1B0GIC8 A0A1L8D9L9 A0A1B6DD91 A0A1B6GVD8 A0A1B6L103 A0A2A3ES33 T1GG11 B4PD77 A0A1I8NZ14 A0A1B0GBS6 A0A034V3Z6 D3TNY4 B3NB98 A0A1A9WQM6 A0A0K8U7D0 A0A0C9RXY8 A0A2M4APJ7 A0A2M4BTX1 A0A2M4CUD1 A0A0K8W7F1 A0A2M4BTU3 Q8MSC8 A0A1I8N099 A0A1A9ZB33 B4QM07 A0A0A1WEP1 A0A0K8U4Q7 A0A1A9Y7P0 A0A1B0BEB5 A0A1A9VR60 Q7QH94 B4N3U3 A0A182RD65 A0A182WUD4 A0A182KKT7 A0A182TRT5 A0A182HW41 B4HVS2 A0A182WDS0 A0A182T317 A0A182JYE4 A0A182MME8 A0A182FTL0 A0A182P5C5 A0A182YAU9 A0A182NSX4 A0A182UW35 A0A182QFR8 B0WML1 A0A182ISA6 A0A1Q3FB88 B4H5M1 Q29EW6 Q0IEI7 W8BX70 B4KY06 B3M517 B4LG65 A0A3B0JYW6 A0A182GEN4
A0A1W4W8H8 A0A194PWW6 A0A194QQP5 A0A232EPT5 A0A2J7Q1Z8 K7IM31 A0A310S8H2 E0VIT0 A0A067QZK3 A0A0N0U560 A0A151I5N8 A0A154PNP3 A0A0K8TCI3 A0A146M9Q1 A0A0A9XY89 E9IPK6 A0A195CK58 A0A195EVS3 A0A195DQJ3 A0A158NAJ9 A0A088ANZ2 A0A023F0J5 A0A0L7LJN2 D6WHD1 E2BPJ4 A0A0K8TQP7 E2ABK9 A0A069DRD9 A0A151X5E4 A0A026WWR9 A0A1B6HX94 A0A0L7QN51 A0A0V0GDD1 A0A1Y1K228 A0A1B0GIC8 A0A1L8D9L9 A0A1B6DD91 A0A1B6GVD8 A0A1B6L103 A0A2A3ES33 T1GG11 B4PD77 A0A1I8NZ14 A0A1B0GBS6 A0A034V3Z6 D3TNY4 B3NB98 A0A1A9WQM6 A0A0K8U7D0 A0A0C9RXY8 A0A2M4APJ7 A0A2M4BTX1 A0A2M4CUD1 A0A0K8W7F1 A0A2M4BTU3 Q8MSC8 A0A1I8N099 A0A1A9ZB33 B4QM07 A0A0A1WEP1 A0A0K8U4Q7 A0A1A9Y7P0 A0A1B0BEB5 A0A1A9VR60 Q7QH94 B4N3U3 A0A182RD65 A0A182WUD4 A0A182KKT7 A0A182TRT5 A0A182HW41 B4HVS2 A0A182WDS0 A0A182T317 A0A182JYE4 A0A182MME8 A0A182FTL0 A0A182P5C5 A0A182YAU9 A0A182NSX4 A0A182UW35 A0A182QFR8 B0WML1 A0A182ISA6 A0A1Q3FB88 B4H5M1 Q29EW6 Q0IEI7 W8BX70 B4KY06 B3M517 B4LG65 A0A3B0JYW6 A0A182GEN4
Pubmed
19121390
23622113
22118469
26354079
28648823
20075255
+ More
20566863 24845553 26823975 25401762 21282665 21347285 25474469 26227816 18362917 19820115 20798317 26369729 26334808 24508170 30249741 28004739 17994087 17550304 25348373 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 22936249 25830018 12364791 20966253 25244985 15632085 17510324 24495485 26483478
20566863 24845553 26823975 25401762 21282665 21347285 25474469 26227816 18362917 19820115 20798317 26369729 26334808 24508170 30249741 28004739 17994087 17550304 25348373 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 22936249 25830018 12364791 20966253 25244985 15632085 17510324 24495485 26483478
EMBL
BABH01043858
NWSH01002655
PCG67749.1
ODYU01005896
SOQ47256.1
RSAL01000043
+ More
RVE50727.1 GAIX01013878 JAA78682.1 AGBW02003612 OWR55531.1 KQ459590 KPI97254.1 KQ461190 KPJ07290.1 NNAY01002909 OXU20342.1 NEVH01019376 PNF22589.1 KQ770827 OAD52581.1 AAZO01002694 AAZO01002695 DS235205 EEB13286.1 KK852855 KDR14945.1 KQ435796 KOX73544.1 KQ976413 KYM90391.1 KQ435007 KZC13473.1 GBRD01002710 JAG63111.1 GDHC01002151 JAQ16478.1 GBHO01019776 GBHO01019773 JAG23828.1 JAG23831.1 GL764545 EFZ17497.1 KQ977642 KYN01106.1 KQ981954 KYN32241.1 KQ980612 KYN15133.1 ADTU01010342 GBBI01004206 JAC14506.1 JTDY01000862 KOB75635.1 KQ971321 EFA00643.1 GL449633 EFN82398.1 GDAI01001125 JAI16478.1 GL438297 EFN69180.1 GBGD01002677 JAC86212.1 KQ982519 KYQ55554.1 KK107077 QOIP01000010 EZA60525.1 RLU17720.1 GECU01028419 JAS79287.1 KQ414860 KOC60055.1 GECL01000271 JAP05853.1 GEZM01095059 JAV55532.1 AJWK01013684 GFDF01010923 JAV03161.1 GEDC01013743 JAS23555.1 GECZ01003402 JAS66367.1 GEBQ01022604 JAT17373.1 KZ288193 PBC34026.1 CAQQ02049520 CM000159 EDW92825.1 CCAG010005888 GAKP01022457 JAC36495.1 EZ423136 ADD19412.1 CH954178 EDV50151.1 GDHF01029821 JAI22493.1 GBYB01012751 JAG82518.1 GGFK01009207 MBW42528.1 GGFJ01007358 MBW56499.1 GGFL01004745 MBW68923.1 GDHF01005335 JAI46979.1 GGFJ01007359 MBW56500.1 AE014296 AY118903 AAF47483.3 AAM50763.1 CM000363 CM002912 EDX08793.1 KMY96805.1 GBXI01017131 JAC97160.1 GDHF01031044 JAI21270.1 JXJN01012834 AAAB01008816 EAA05078.4 CH964095 EDW79298.1 APCN01001462 CH480817 EDW50037.1 AXCM01003024 AXCN02002134 DS231998 EDS31082.1 GFDL01010225 JAV24820.1 CH479211 EDW33073.1 CH379070 EAL29943.2 CH477655 EAT37723.1 GAMC01005117 GAMC01005116 JAC01439.1 CH933809 EDW18708.1 CH902618 EDV39496.1 CH940647 EDW69373.1 OUUW01000012 SPP87265.1 JXUM01011718 KQ560337 KXJ82971.1
RVE50727.1 GAIX01013878 JAA78682.1 AGBW02003612 OWR55531.1 KQ459590 KPI97254.1 KQ461190 KPJ07290.1 NNAY01002909 OXU20342.1 NEVH01019376 PNF22589.1 KQ770827 OAD52581.1 AAZO01002694 AAZO01002695 DS235205 EEB13286.1 KK852855 KDR14945.1 KQ435796 KOX73544.1 KQ976413 KYM90391.1 KQ435007 KZC13473.1 GBRD01002710 JAG63111.1 GDHC01002151 JAQ16478.1 GBHO01019776 GBHO01019773 JAG23828.1 JAG23831.1 GL764545 EFZ17497.1 KQ977642 KYN01106.1 KQ981954 KYN32241.1 KQ980612 KYN15133.1 ADTU01010342 GBBI01004206 JAC14506.1 JTDY01000862 KOB75635.1 KQ971321 EFA00643.1 GL449633 EFN82398.1 GDAI01001125 JAI16478.1 GL438297 EFN69180.1 GBGD01002677 JAC86212.1 KQ982519 KYQ55554.1 KK107077 QOIP01000010 EZA60525.1 RLU17720.1 GECU01028419 JAS79287.1 KQ414860 KOC60055.1 GECL01000271 JAP05853.1 GEZM01095059 JAV55532.1 AJWK01013684 GFDF01010923 JAV03161.1 GEDC01013743 JAS23555.1 GECZ01003402 JAS66367.1 GEBQ01022604 JAT17373.1 KZ288193 PBC34026.1 CAQQ02049520 CM000159 EDW92825.1 CCAG010005888 GAKP01022457 JAC36495.1 EZ423136 ADD19412.1 CH954178 EDV50151.1 GDHF01029821 JAI22493.1 GBYB01012751 JAG82518.1 GGFK01009207 MBW42528.1 GGFJ01007358 MBW56499.1 GGFL01004745 MBW68923.1 GDHF01005335 JAI46979.1 GGFJ01007359 MBW56500.1 AE014296 AY118903 AAF47483.3 AAM50763.1 CM000363 CM002912 EDX08793.1 KMY96805.1 GBXI01017131 JAC97160.1 GDHF01031044 JAI21270.1 JXJN01012834 AAAB01008816 EAA05078.4 CH964095 EDW79298.1 APCN01001462 CH480817 EDW50037.1 AXCM01003024 AXCN02002134 DS231998 EDS31082.1 GFDL01010225 JAV24820.1 CH479211 EDW33073.1 CH379070 EAL29943.2 CH477655 EAT37723.1 GAMC01005117 GAMC01005116 JAC01439.1 CH933809 EDW18708.1 CH902618 EDV39496.1 CH940647 EDW69373.1 OUUW01000012 SPP87265.1 JXUM01011718 KQ560337 KXJ82971.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000192223
UP000053268
+ More
UP000053240 UP000215335 UP000235965 UP000002358 UP000009046 UP000027135 UP000053105 UP000078540 UP000076502 UP000078542 UP000078541 UP000078492 UP000005205 UP000005203 UP000037510 UP000007266 UP000008237 UP000000311 UP000075809 UP000053097 UP000279307 UP000053825 UP000092461 UP000242457 UP000015102 UP000002282 UP000095300 UP000092444 UP000008711 UP000091820 UP000000803 UP000095301 UP000092445 UP000000304 UP000092443 UP000092460 UP000078200 UP000007062 UP000007798 UP000075900 UP000076407 UP000075882 UP000075902 UP000075840 UP000001292 UP000075920 UP000075901 UP000075881 UP000075883 UP000069272 UP000075885 UP000076408 UP000075884 UP000075903 UP000075886 UP000002320 UP000075880 UP000008744 UP000001819 UP000008820 UP000009192 UP000007801 UP000008792 UP000268350 UP000069940 UP000249989
UP000053240 UP000215335 UP000235965 UP000002358 UP000009046 UP000027135 UP000053105 UP000078540 UP000076502 UP000078542 UP000078541 UP000078492 UP000005205 UP000005203 UP000037510 UP000007266 UP000008237 UP000000311 UP000075809 UP000053097 UP000279307 UP000053825 UP000092461 UP000242457 UP000015102 UP000002282 UP000095300 UP000092444 UP000008711 UP000091820 UP000000803 UP000095301 UP000092445 UP000000304 UP000092443 UP000092460 UP000078200 UP000007062 UP000007798 UP000075900 UP000076407 UP000075882 UP000075902 UP000075840 UP000001292 UP000075920 UP000075901 UP000075881 UP000075883 UP000069272 UP000075885 UP000076408 UP000075884 UP000075903 UP000075886 UP000002320 UP000075880 UP000008744 UP000001819 UP000008820 UP000009192 UP000007801 UP000008792 UP000268350 UP000069940 UP000249989
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9JYI6
A0A2A4J797
A0A2H1W2T7
A0A3S2LCM1
S4PSF5
A0A212FP60
+ More
A0A1W4W8H8 A0A194PWW6 A0A194QQP5 A0A232EPT5 A0A2J7Q1Z8 K7IM31 A0A310S8H2 E0VIT0 A0A067QZK3 A0A0N0U560 A0A151I5N8 A0A154PNP3 A0A0K8TCI3 A0A146M9Q1 A0A0A9XY89 E9IPK6 A0A195CK58 A0A195EVS3 A0A195DQJ3 A0A158NAJ9 A0A088ANZ2 A0A023F0J5 A0A0L7LJN2 D6WHD1 E2BPJ4 A0A0K8TQP7 E2ABK9 A0A069DRD9 A0A151X5E4 A0A026WWR9 A0A1B6HX94 A0A0L7QN51 A0A0V0GDD1 A0A1Y1K228 A0A1B0GIC8 A0A1L8D9L9 A0A1B6DD91 A0A1B6GVD8 A0A1B6L103 A0A2A3ES33 T1GG11 B4PD77 A0A1I8NZ14 A0A1B0GBS6 A0A034V3Z6 D3TNY4 B3NB98 A0A1A9WQM6 A0A0K8U7D0 A0A0C9RXY8 A0A2M4APJ7 A0A2M4BTX1 A0A2M4CUD1 A0A0K8W7F1 A0A2M4BTU3 Q8MSC8 A0A1I8N099 A0A1A9ZB33 B4QM07 A0A0A1WEP1 A0A0K8U4Q7 A0A1A9Y7P0 A0A1B0BEB5 A0A1A9VR60 Q7QH94 B4N3U3 A0A182RD65 A0A182WUD4 A0A182KKT7 A0A182TRT5 A0A182HW41 B4HVS2 A0A182WDS0 A0A182T317 A0A182JYE4 A0A182MME8 A0A182FTL0 A0A182P5C5 A0A182YAU9 A0A182NSX4 A0A182UW35 A0A182QFR8 B0WML1 A0A182ISA6 A0A1Q3FB88 B4H5M1 Q29EW6 Q0IEI7 W8BX70 B4KY06 B3M517 B4LG65 A0A3B0JYW6 A0A182GEN4
A0A1W4W8H8 A0A194PWW6 A0A194QQP5 A0A232EPT5 A0A2J7Q1Z8 K7IM31 A0A310S8H2 E0VIT0 A0A067QZK3 A0A0N0U560 A0A151I5N8 A0A154PNP3 A0A0K8TCI3 A0A146M9Q1 A0A0A9XY89 E9IPK6 A0A195CK58 A0A195EVS3 A0A195DQJ3 A0A158NAJ9 A0A088ANZ2 A0A023F0J5 A0A0L7LJN2 D6WHD1 E2BPJ4 A0A0K8TQP7 E2ABK9 A0A069DRD9 A0A151X5E4 A0A026WWR9 A0A1B6HX94 A0A0L7QN51 A0A0V0GDD1 A0A1Y1K228 A0A1B0GIC8 A0A1L8D9L9 A0A1B6DD91 A0A1B6GVD8 A0A1B6L103 A0A2A3ES33 T1GG11 B4PD77 A0A1I8NZ14 A0A1B0GBS6 A0A034V3Z6 D3TNY4 B3NB98 A0A1A9WQM6 A0A0K8U7D0 A0A0C9RXY8 A0A2M4APJ7 A0A2M4BTX1 A0A2M4CUD1 A0A0K8W7F1 A0A2M4BTU3 Q8MSC8 A0A1I8N099 A0A1A9ZB33 B4QM07 A0A0A1WEP1 A0A0K8U4Q7 A0A1A9Y7P0 A0A1B0BEB5 A0A1A9VR60 Q7QH94 B4N3U3 A0A182RD65 A0A182WUD4 A0A182KKT7 A0A182TRT5 A0A182HW41 B4HVS2 A0A182WDS0 A0A182T317 A0A182JYE4 A0A182MME8 A0A182FTL0 A0A182P5C5 A0A182YAU9 A0A182NSX4 A0A182UW35 A0A182QFR8 B0WML1 A0A182ISA6 A0A1Q3FB88 B4H5M1 Q29EW6 Q0IEI7 W8BX70 B4KY06 B3M517 B4LG65 A0A3B0JYW6 A0A182GEN4
Ontologies
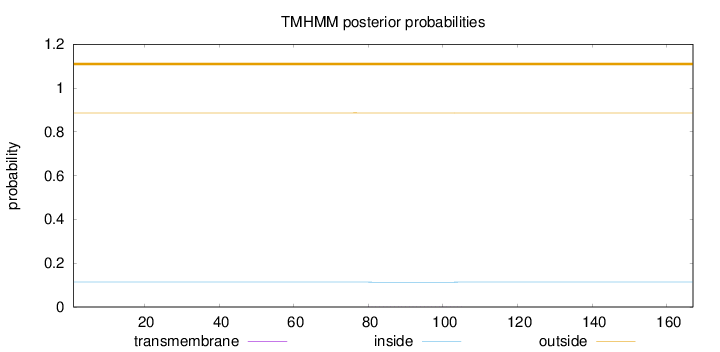
Topology
Length:
167
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01334
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11364
outside
1 - 167
Population Genetic Test Statistics
Pi
52.572207
Theta
64.792088
Tajima's D
-0.588836
CLR
101.789513
CSRT
0.218839058047098
Interpretation
Uncertain
