Gene
KWMTBOMO06525
Pre Gene Modal
BGIBMGA011643
Annotation
Methyltransferase-like_protein_22_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.772 PlasmaMembrane Reliability : 1.772
Sequence
CDS
ATGCCTGAGTACACAGTAACATCAGAGATTTACGAAGAATATGATTACACAAGTAATATAACTCCCAGCGCAAATGGAAATGTCATATCGGAATTTCCATTTTTGCTACCCATAACCGGACAAACTGCTAAGTTTGATGAAGACGGTGACCTAGATGCTGTAAGACCTAAAAGAAAAGTTATAAGAATTGAACACAGCCCCAAAACGGCAATATCTTTAGTGGGCCGACAAGTATGGAGAGGAGCATTTCTATTAGGAGACTGGTTAATACACCTTGGCTTAAAAGGCGAGCTCTCCAATAAGAATGTTCTAGAATTAGGAGCAGGTACAGGAGTGACGAGTATCGTCGCTGCCCTGTTCGCTGAGAAAGTAGTGTGTACAGACATCAACGTAGGAGGTATCCTGGATCTAATAAACCTGAATGCGAAGTACAATAAGGATTTAATAAAATCACATTTCTCTGTAATGCCGCTGGACTTCACCGATACCGCGTGGAGTTCAAAACTTAATAATGTGATCAAAGAAGTCGACTTCATTATTGCGGCTGATGTGATATACGACGATGATGTGACAGCAGCCTTCATATCCACTATACAAAAGATACTTAACTTGAAACCTCCAAAAACTGTGTACGTTGTGTTGGAGAAGCGTTATGTGTTTACAATAGAACATCTGGATTGCGTCGCGCCGTGTTACGAGACATTCCTCACTTTGTTGGACAAAGTGAAATCAGAAAACGTTCATTCCACTTGGACAGTCGAACAACTACCTCTAGATTTTCCAAAATATTTCACTTACGACAGAGTGAAAGATCTAGTGTTATGGAAGATATCATCCAAATAG
Protein
MPEYTVTSEIYEEYDYTSNITPSANGNVISEFPFLLPITGQTAKFDEDGDLDAVRPKRKVIRIEHSPKTAISLVGRQVWRGAFLLGDWLIHLGLKGELSNKNVLELGAGTGVTSIVAALFAEKVVCTDINVGGILDLINLNAKYNKDLIKSHFSVMPLDFTDTAWSSKLNNVIKEVDFIIAADVIYDDDVTAAFISTIQKILNLKPPKTVYVVLEKRYVFTIEHLDCVAPCYETFLTLLDKVKSENVHSTWTVEQLPLDFPKYFTYDRVKDLVLWKISSK
Summary
Uniprot
H9JQ37
A0A2A4J8L4
A0A2W1BL16
A0A0L7LJA5
A0A194PV25
A0A194QP39
+ More
A0A212FP55 S4PTF6 A0A2J7R345 A0A067RKT2 A0A1B6GD54 A0A1B6LBI9 A0A2J7R327 A0A0M4EZ50 E2B287 A0A151IEN4 B4LHG3 B4KWM7 U4UQB3 A0A1W4WIU4 A0A1W4VAT0 N6T0J9 T1GBZ4 B4QJM0 A0A0A9X8Z4 Q2M161 E0VGK4 Q9VP86 B3NIV0 B4IAJ0 A0A1Y1KSM1 A0A232FJB1 A0A3B0JA90 B4J082 B4IUK1 A0A151XCS1 B3M6U8 A0A336MR32 A0A336MUV3 A0A1I8PT16 B4N3V3 B4PE81 A0A0P4VV39 K7IM37 A0A1A9X0M8 D6WHD5 A0A1I8NFE7 W8C3Z1 A0A1I8MCU9 A0A1A9XJG3 A0A1B0AY75 A0A3L8DCG1 A0A0L0CNM4 A0A0K8VHP3 A0A1B0FBM2 A0A026VWS1 E2BG04 A0A182RRZ1 A0A182Y9E5 A0A195FI98 A0A195AVP1 A0A1B0A6I7 A0A195DMK7 A0A1B0DJ88 A0A158P234 A0A182M5C7 A0A034WLC7 A0A1A9UJ69 A0A0K8TGH5 A0A182WDX3 A0A2J7R334 A0A2J7R326 A0A182Q1J3 A0A084WIP8 F4W6B5 A0A146MEB7 A0A0L7QZH8 A0A182JI46 A0A2M4BSK8 A0A310SCJ9 A0A182JRA5 Q7PZF2 A0A182FB41 A0A0M8ZNT1 A0A182VA20 A0A182U3X5 A0A3Q0JLP5 A0A1S4H7S5 A0A182HHH7 A0A1B0CV25 A0A2M3Z9U1 A0A3F2YUS4 A0A182XJV6 A0A087YU40 A0A1Q3FGU5 A0A2M4AU92 A0A182N3Z7 Q17FA6
A0A212FP55 S4PTF6 A0A2J7R345 A0A067RKT2 A0A1B6GD54 A0A1B6LBI9 A0A2J7R327 A0A0M4EZ50 E2B287 A0A151IEN4 B4LHG3 B4KWM7 U4UQB3 A0A1W4WIU4 A0A1W4VAT0 N6T0J9 T1GBZ4 B4QJM0 A0A0A9X8Z4 Q2M161 E0VGK4 Q9VP86 B3NIV0 B4IAJ0 A0A1Y1KSM1 A0A232FJB1 A0A3B0JA90 B4J082 B4IUK1 A0A151XCS1 B3M6U8 A0A336MR32 A0A336MUV3 A0A1I8PT16 B4N3V3 B4PE81 A0A0P4VV39 K7IM37 A0A1A9X0M8 D6WHD5 A0A1I8NFE7 W8C3Z1 A0A1I8MCU9 A0A1A9XJG3 A0A1B0AY75 A0A3L8DCG1 A0A0L0CNM4 A0A0K8VHP3 A0A1B0FBM2 A0A026VWS1 E2BG04 A0A182RRZ1 A0A182Y9E5 A0A195FI98 A0A195AVP1 A0A1B0A6I7 A0A195DMK7 A0A1B0DJ88 A0A158P234 A0A182M5C7 A0A034WLC7 A0A1A9UJ69 A0A0K8TGH5 A0A182WDX3 A0A2J7R334 A0A2J7R326 A0A182Q1J3 A0A084WIP8 F4W6B5 A0A146MEB7 A0A0L7QZH8 A0A182JI46 A0A2M4BSK8 A0A310SCJ9 A0A182JRA5 Q7PZF2 A0A182FB41 A0A0M8ZNT1 A0A182VA20 A0A182U3X5 A0A3Q0JLP5 A0A1S4H7S5 A0A182HHH7 A0A1B0CV25 A0A2M3Z9U1 A0A3F2YUS4 A0A182XJV6 A0A087YU40 A0A1Q3FGU5 A0A2M4AU92 A0A182N3Z7 Q17FA6
Pubmed
19121390
28756777
26227816
26354079
22118469
23622113
+ More
24845553 20798317 17994087 23537049 22936249 25401762 15632085 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 28648823 17550304 20075255 18362917 19820115 25315136 24495485 30249741 26108605 24508170 25244985 21347285 25348373 24438588 21719571 26823975 12364791 20966253 20920257 17510324
24845553 20798317 17994087 23537049 22936249 25401762 15632085 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 28648823 17550304 20075255 18362917 19820115 25315136 24495485 30249741 26108605 24508170 25244985 21347285 25348373 24438588 21719571 26823975 12364791 20966253 20920257 17510324
EMBL
BABH01012177
NWSH01002655
PCG67752.1
KZ149984
PZC75729.1
JTDY01000862
+ More
KOB75633.1 KQ459590 KPI97251.1 KQ461190 KPJ07293.1 AGBW02003612 OWR55528.1 GAIX01012123 JAA80437.1 NEVH01007823 PNF35246.1 KK852567 KDR21190.1 GECZ01030370 GECZ01028981 GECZ01009418 GECZ01008540 GECZ01000357 JAS39399.1 JAS40788.1 JAS60351.1 JAS61229.1 JAS69412.1 GEBQ01019083 JAT20894.1 PNF35249.1 CP012525 ALC43607.1 GL445067 EFN60213.1 KQ977858 KYM99299.1 CH940647 EDW68493.1 CH933809 EDW17474.1 KB632417 ERL95287.1 APGK01049655 KB741156 KB630646 KB631725 ENN73589.1 ERL83763.1 ERL85607.1 CAQQ02076762 CM000363 CM002912 EDX11320.1 KMZ00917.1 GBHO01030047 GBHO01030046 GBHO01030044 GBHO01026478 GBRD01007212 GBRD01007208 JAG13557.1 JAG13558.1 JAG13560.1 JAG17126.1 JAG58609.1 CH379069 EAL30714.1 DS235148 EEB12510.1 AE014296 BT029075 AAF51669.1 ABJ17008.1 CH954178 EDV52596.1 CH480826 EDW44303.1 GEZM01077735 JAV63190.1 NNAY01000146 OXU30569.1 OUUW01000002 SPP76902.1 CH916366 EDV95683.1 CH891933 EDX00065.2 KQ982297 KYQ58176.1 CH902618 EDV39784.1 UFQT01002105 SSX32696.1 UFQS01002193 UFQT01002193 SSX13458.1 SSX32889.1 CH964095 EDW79308.1 CM000159 EDW95094.2 GDRN01094582 JAI59705.1 KQ971321 EFA00105.1 GAMC01002572 JAC03984.1 JXJN01005555 QOIP01000010 RLU18044.1 JRES01000128 KNC33938.1 GDHF01013908 JAI38406.1 CCAG010008960 KK107670 EZA48187.1 GL448096 EFN85406.1 KQ981523 KYN40400.1 KQ976731 KYM76246.1 KQ980724 KYN14066.1 AJVK01034893 ADTU01006974 ADTU01006975 AXCM01001557 GAKP01003493 GAKP01003491 JAC55459.1 GBRD01007209 GBRD01001128 JAG64693.1 PNF35248.1 PNF35247.1 AXCN02001090 ATLV01023941 KE525347 KFB50092.1 GL887707 EGI70250.1 GDHC01001543 JAQ17086.1 KQ414681 KOC63947.1 GGFJ01006627 MBW55768.1 KQ761402 OAD57570.1 AAAB01008986 EAA00499.4 KQ435955 KOX68070.1 APCN01002416 AJWK01030034 GGFM01004543 MBW25294.1 GGFL01000971 MBW65149.1 GFDL01008303 JAV26742.1 GGFK01011039 MBW44360.1 CH477272 EAT45241.1 EJY57487.1
KOB75633.1 KQ459590 KPI97251.1 KQ461190 KPJ07293.1 AGBW02003612 OWR55528.1 GAIX01012123 JAA80437.1 NEVH01007823 PNF35246.1 KK852567 KDR21190.1 GECZ01030370 GECZ01028981 GECZ01009418 GECZ01008540 GECZ01000357 JAS39399.1 JAS40788.1 JAS60351.1 JAS61229.1 JAS69412.1 GEBQ01019083 JAT20894.1 PNF35249.1 CP012525 ALC43607.1 GL445067 EFN60213.1 KQ977858 KYM99299.1 CH940647 EDW68493.1 CH933809 EDW17474.1 KB632417 ERL95287.1 APGK01049655 KB741156 KB630646 KB631725 ENN73589.1 ERL83763.1 ERL85607.1 CAQQ02076762 CM000363 CM002912 EDX11320.1 KMZ00917.1 GBHO01030047 GBHO01030046 GBHO01030044 GBHO01026478 GBRD01007212 GBRD01007208 JAG13557.1 JAG13558.1 JAG13560.1 JAG17126.1 JAG58609.1 CH379069 EAL30714.1 DS235148 EEB12510.1 AE014296 BT029075 AAF51669.1 ABJ17008.1 CH954178 EDV52596.1 CH480826 EDW44303.1 GEZM01077735 JAV63190.1 NNAY01000146 OXU30569.1 OUUW01000002 SPP76902.1 CH916366 EDV95683.1 CH891933 EDX00065.2 KQ982297 KYQ58176.1 CH902618 EDV39784.1 UFQT01002105 SSX32696.1 UFQS01002193 UFQT01002193 SSX13458.1 SSX32889.1 CH964095 EDW79308.1 CM000159 EDW95094.2 GDRN01094582 JAI59705.1 KQ971321 EFA00105.1 GAMC01002572 JAC03984.1 JXJN01005555 QOIP01000010 RLU18044.1 JRES01000128 KNC33938.1 GDHF01013908 JAI38406.1 CCAG010008960 KK107670 EZA48187.1 GL448096 EFN85406.1 KQ981523 KYN40400.1 KQ976731 KYM76246.1 KQ980724 KYN14066.1 AJVK01034893 ADTU01006974 ADTU01006975 AXCM01001557 GAKP01003493 GAKP01003491 JAC55459.1 GBRD01007209 GBRD01001128 JAG64693.1 PNF35248.1 PNF35247.1 AXCN02001090 ATLV01023941 KE525347 KFB50092.1 GL887707 EGI70250.1 GDHC01001543 JAQ17086.1 KQ414681 KOC63947.1 GGFJ01006627 MBW55768.1 KQ761402 OAD57570.1 AAAB01008986 EAA00499.4 KQ435955 KOX68070.1 APCN01002416 AJWK01030034 GGFM01004543 MBW25294.1 GGFL01000971 MBW65149.1 GFDL01008303 JAV26742.1 GGFK01011039 MBW44360.1 CH477272 EAT45241.1 EJY57487.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000007151
+ More
UP000235965 UP000027135 UP000092553 UP000000311 UP000078542 UP000008792 UP000009192 UP000030742 UP000192223 UP000192221 UP000019118 UP000015102 UP000000304 UP000001819 UP000009046 UP000000803 UP000008711 UP000001292 UP000215335 UP000268350 UP000001070 UP000002282 UP000075809 UP000007801 UP000095300 UP000007798 UP000002358 UP000091820 UP000007266 UP000095301 UP000092443 UP000092460 UP000279307 UP000037069 UP000092444 UP000053097 UP000008237 UP000075900 UP000076408 UP000078541 UP000078540 UP000092445 UP000078492 UP000092462 UP000005205 UP000075883 UP000078200 UP000075920 UP000075886 UP000030765 UP000007755 UP000053825 UP000075880 UP000075881 UP000007062 UP000069272 UP000053105 UP000075903 UP000075902 UP000079169 UP000075840 UP000092461 UP000075882 UP000076407 UP000075884 UP000008820
UP000235965 UP000027135 UP000092553 UP000000311 UP000078542 UP000008792 UP000009192 UP000030742 UP000192223 UP000192221 UP000019118 UP000015102 UP000000304 UP000001819 UP000009046 UP000000803 UP000008711 UP000001292 UP000215335 UP000268350 UP000001070 UP000002282 UP000075809 UP000007801 UP000095300 UP000007798 UP000002358 UP000091820 UP000007266 UP000095301 UP000092443 UP000092460 UP000279307 UP000037069 UP000092444 UP000053097 UP000008237 UP000075900 UP000076408 UP000078541 UP000078540 UP000092445 UP000078492 UP000092462 UP000005205 UP000075883 UP000078200 UP000075920 UP000075886 UP000030765 UP000007755 UP000053825 UP000075880 UP000075881 UP000007062 UP000069272 UP000053105 UP000075903 UP000075902 UP000079169 UP000075840 UP000092461 UP000075882 UP000076407 UP000075884 UP000008820
Pfam
PF10294 Methyltransf_16
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9JQ37
A0A2A4J8L4
A0A2W1BL16
A0A0L7LJA5
A0A194PV25
A0A194QP39
+ More
A0A212FP55 S4PTF6 A0A2J7R345 A0A067RKT2 A0A1B6GD54 A0A1B6LBI9 A0A2J7R327 A0A0M4EZ50 E2B287 A0A151IEN4 B4LHG3 B4KWM7 U4UQB3 A0A1W4WIU4 A0A1W4VAT0 N6T0J9 T1GBZ4 B4QJM0 A0A0A9X8Z4 Q2M161 E0VGK4 Q9VP86 B3NIV0 B4IAJ0 A0A1Y1KSM1 A0A232FJB1 A0A3B0JA90 B4J082 B4IUK1 A0A151XCS1 B3M6U8 A0A336MR32 A0A336MUV3 A0A1I8PT16 B4N3V3 B4PE81 A0A0P4VV39 K7IM37 A0A1A9X0M8 D6WHD5 A0A1I8NFE7 W8C3Z1 A0A1I8MCU9 A0A1A9XJG3 A0A1B0AY75 A0A3L8DCG1 A0A0L0CNM4 A0A0K8VHP3 A0A1B0FBM2 A0A026VWS1 E2BG04 A0A182RRZ1 A0A182Y9E5 A0A195FI98 A0A195AVP1 A0A1B0A6I7 A0A195DMK7 A0A1B0DJ88 A0A158P234 A0A182M5C7 A0A034WLC7 A0A1A9UJ69 A0A0K8TGH5 A0A182WDX3 A0A2J7R334 A0A2J7R326 A0A182Q1J3 A0A084WIP8 F4W6B5 A0A146MEB7 A0A0L7QZH8 A0A182JI46 A0A2M4BSK8 A0A310SCJ9 A0A182JRA5 Q7PZF2 A0A182FB41 A0A0M8ZNT1 A0A182VA20 A0A182U3X5 A0A3Q0JLP5 A0A1S4H7S5 A0A182HHH7 A0A1B0CV25 A0A2M3Z9U1 A0A3F2YUS4 A0A182XJV6 A0A087YU40 A0A1Q3FGU5 A0A2M4AU92 A0A182N3Z7 Q17FA6
A0A212FP55 S4PTF6 A0A2J7R345 A0A067RKT2 A0A1B6GD54 A0A1B6LBI9 A0A2J7R327 A0A0M4EZ50 E2B287 A0A151IEN4 B4LHG3 B4KWM7 U4UQB3 A0A1W4WIU4 A0A1W4VAT0 N6T0J9 T1GBZ4 B4QJM0 A0A0A9X8Z4 Q2M161 E0VGK4 Q9VP86 B3NIV0 B4IAJ0 A0A1Y1KSM1 A0A232FJB1 A0A3B0JA90 B4J082 B4IUK1 A0A151XCS1 B3M6U8 A0A336MR32 A0A336MUV3 A0A1I8PT16 B4N3V3 B4PE81 A0A0P4VV39 K7IM37 A0A1A9X0M8 D6WHD5 A0A1I8NFE7 W8C3Z1 A0A1I8MCU9 A0A1A9XJG3 A0A1B0AY75 A0A3L8DCG1 A0A0L0CNM4 A0A0K8VHP3 A0A1B0FBM2 A0A026VWS1 E2BG04 A0A182RRZ1 A0A182Y9E5 A0A195FI98 A0A195AVP1 A0A1B0A6I7 A0A195DMK7 A0A1B0DJ88 A0A158P234 A0A182M5C7 A0A034WLC7 A0A1A9UJ69 A0A0K8TGH5 A0A182WDX3 A0A2J7R334 A0A2J7R326 A0A182Q1J3 A0A084WIP8 F4W6B5 A0A146MEB7 A0A0L7QZH8 A0A182JI46 A0A2M4BSK8 A0A310SCJ9 A0A182JRA5 Q7PZF2 A0A182FB41 A0A0M8ZNT1 A0A182VA20 A0A182U3X5 A0A3Q0JLP5 A0A1S4H7S5 A0A182HHH7 A0A1B0CV25 A0A2M3Z9U1 A0A3F2YUS4 A0A182XJV6 A0A087YU40 A0A1Q3FGU5 A0A2M4AU92 A0A182N3Z7 Q17FA6
PDB
4LG1
E-value=1.42837e-05,
Score=114
Ontologies
PANTHER
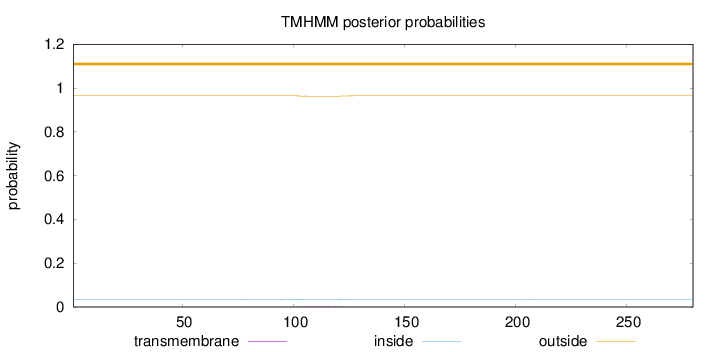
Topology
Length:
280
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0987400000000002
Exp number, first 60 AAs:
0.00412
Total prob of N-in:
0.03549
outside
1 - 280
Population Genetic Test Statistics
Pi
319.717444
Theta
181.111358
Tajima's D
1.6377
CLR
0.23827
CSRT
0.821608919554022
Interpretation
Uncertain
