Gene
KWMTBOMO06524 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011844
Annotation
protein_disulfide_isomerase_precursor_[Bombyx_mori]
Full name
Protein disulfide-isomerase
Location in the cell
ER Reliability : 4.553
Sequence
CDS
ATGCGTGTTTTAATTTTCACGGCTATAGCCCTGCTAGGGCTCGCTCTCGGAGATGAAGTGCCCACTGAAGAAAATGTGCTCGTTTTGAGTAAAGCTAACTTTGAAACTGTAATTACAACCACGGAGTACATTTTAGTTGAATTCTATGCTCCATGGTGCGGCCACTGCAAATCTCTGGCACCGGAATACGCCAAGGCAGCAACAAAGCTGGCTGAAGAAGAATCTCCTATCAAACTAGCGAAAGTTGACGCAACTCAAGAACAGGATCTCGCCGAGAGCTACGGTGTACGAGGATACCCGACTCTCAAATTCTTCAGGAATGGCAGTCCTATCGACTATTCAGGTGGTCGTCAAGCTGATGACATCATCAGTTGGCTGAAGAAGAAGACTGGCCCCCCTGCTGTTGAGGTCACCTCTGCTGAACAGGCTAAAGAACTTATCGATGCCAATACTGTTATTGTATTTGGTTTCTTTTCGGACCAGAGCTCAGCCAGAGCCAAAACTTTCCTTTCAACTGCTCAAGTTGTTGATGACCAAGTATTTGCTATTGTCAGCGATGAGAAAGTGATCAAGGAGTTGGAGGCTGAAGATGAAGATGTTGTGCTTTTCAAGAACTTCGAAGAGAAACGTGTCAAATATGAAGATGAAGAAATCACTGAGGATCTGCTCAATGCTTGGGTGTTTGTCCAGAGCATGCCTACTATCGTTGAATTCTCGCATGAAACTGCTTCCAAGATCTTTGGTGGAAAGATCAAATACCACCTTCTTATCTTCTTATCCAAGAAAAATGGCGATTTCGAGAAATACCTTGAGGATTTGAAGCCGGTCGCGAAGACCTACCGCGACAGGATCATGACCGTCGCCATCGATGCCGACGAAGATGAACACCAAAGGATCTTGGAGTTCTTCGGCATGAAGAAGGATGAGGTTCCATCTGCCCGTCTGATCGCCCTTGAACAAGACATGGCCAAATACAAGCCCAGCAGTAATGAGCTGTCGCCTAACGCTATTGAGGAATTCGTTCAATCGTTCTTCGACGGCACCCTGAAGCAGCATCTGCTGAGCGAGGACCTGCCCTCGGACTGGGCCGCCAAACCCGTCAAAGTGCTCGTCGCCGCCAACTTCGACGAAGTCGTCTTCGACACAACTAAGAAGGTCCTCGTCGAATTCTACGCTCCTTGGTGCGGCCACTGCAAACAGCTGGTCCCTATCTACGACAAGCTCGGCGAGCACTTTGAGAACGACGATGACGTCATCATTGCCAAAATCGACGCCACCGCCAACGAACTGGAACACACCAAGATCACGTCCTTCCCAACGATCAAACTTTACAGCAAGGACAACCAGGTCCACGACTACAACGGCGAGAGGACACTGGCCGGCCTCACCAAGTTCGTTGAGACCGACGGTGAAGGCGCCGAGCCAGGACCTTCTGTGACCGAATTCGAAGAGGAGGAAGATGTGCCTGCCAGAGACGAGTTATAA
Protein
MRVLIFTAIALLGLALGDEVPTEENVLVLSKANFETVITTTEYILVEFYAPWCGHCKSLAPEYAKAATKLAEEESPIKLAKVDATQEQDLAESYGVRGYPTLKFFRNGSPIDYSGGRQADDIISWLKKKTGPPAVEVTSAEQAKELIDANTVIVFGFFSDQSSARAKTFLSTAQVVDDQVFAIVSDEKVIKELEAEDEDVVLFKNFEEKRVKYEDEEITEDLLNAWVFVQSMPTIVEFSHETASKIFGGKIKYHLLIFLSKKNGDFEKYLEDLKPVAKTYRDRIMTVAIDADEDEHQRILEFFGMKKDEVPSARLIALEQDMAKYKPSSNELSPNAIEEFVQSFFDGTLKQHLLSEDLPSDWAAKPVKVLVAANFDEVVFDTTKKVLVEFYAPWCGHCKQLVPIYDKLGEHFENDDDVIIAKIDATANELEHTKITSFPTIKLYSKDNQVHDYNGERTLAGLTKFVETDGEGAEPGPSVTEFEEEEDVPARDEL
Summary
Description
Participates in the folding of proteins containing disulfide bonds.
Catalytic Activity
Catalyzes the rearrangement of -S-S- bonds in proteins.
Subunit
Homodimer.
Similarity
Belongs to the protein disulfide isomerase family.
Keywords
Alternative splicing
Complete proteome
Disulfide bond
Endoplasmic reticulum
Isomerase
Redox-active center
Reference proteome
Repeat
Signal
Feature
chain Protein disulfide-isomerase
splice variant In isoform D.
splice variant In isoform D.
Uniprot
H9JQN8
Q9GPH2
I4DIQ3
A0A2A4J7L8
I4DLY7
A0A0U1VTU3
+ More
A0A2H1VU96 S4NP33 G9D554 A0A212FP73 A0A2W1BSJ9 V5GZ61 A0A1J1I589 A0A1L8DUD1 A0A0P4WI78 A0A1L8DUB0 U5EV80 A0A1L8DUB3 A0A182Y1G6 B0WFQ4 A0A182MYS9 A0A034VQH7 B0X3M7 A0A0K8TWH4 A0A336M5N1 G9JKY3 A0A182TD69 Q5TMX9 A0A1S4HAB3 A0A182KS99 A0A182HQD4 A0A182RD13 A0A182WZT9 A0A182UP09 A0A0P4WLN5 A0A182P5V3 A0A182MG57 A0A1I8MXQ8 A0A2J7R328 A0A1L8EF87 T1PF58 A0A1L8EFE1 A0A182WEX4 W8BHI2 A0A182QYN8 U5EXV6 A0A1I8P5Q9 A0A182J8G7 A0A084WIX0 A0A1L8EFM8 D6WHD6 A0A182FBC4 A0A067RBI3 T1E8D6 A0A2M3ZYI7 A0A2M4BL90 A0A1Q3FFI6 A0A1B6KDW2 A0A2M3ZYQ1 W5JK16 A0A2M3ZYG2 A0A2M4BLF1 A0A1B6IVK8 A0A0A9YM47 A0A1Y1KS02 A0A226ECT4 C0JBY4 R4WQK3 A0A0N8EAP0 A0A182GFX8 Q1HR78 B4L127 A0A3G5ART5 A0A3G5ANU4 B4PIY3 B4QKK6 A0A1W7R9Z2 A0A3B0J7U5 B4HI55 A0A1W4VP72 B3NHN4 T1L2F7 Q2M1A2 X2JGP4 A0A2I9LPF6 P54399 A0A2M3ZE69 B3M733 A0A1Q3FFB3 B4GUE5 E2BG03 B4N522 D3TRE3 B4LGU3 A0A1A9V3N0 B4IXK8 A0A0J7L6A5 A0A2Z5WPU1 A0A0P4VT63
A0A2H1VU96 S4NP33 G9D554 A0A212FP73 A0A2W1BSJ9 V5GZ61 A0A1J1I589 A0A1L8DUD1 A0A0P4WI78 A0A1L8DUB0 U5EV80 A0A1L8DUB3 A0A182Y1G6 B0WFQ4 A0A182MYS9 A0A034VQH7 B0X3M7 A0A0K8TWH4 A0A336M5N1 G9JKY3 A0A182TD69 Q5TMX9 A0A1S4HAB3 A0A182KS99 A0A182HQD4 A0A182RD13 A0A182WZT9 A0A182UP09 A0A0P4WLN5 A0A182P5V3 A0A182MG57 A0A1I8MXQ8 A0A2J7R328 A0A1L8EF87 T1PF58 A0A1L8EFE1 A0A182WEX4 W8BHI2 A0A182QYN8 U5EXV6 A0A1I8P5Q9 A0A182J8G7 A0A084WIX0 A0A1L8EFM8 D6WHD6 A0A182FBC4 A0A067RBI3 T1E8D6 A0A2M3ZYI7 A0A2M4BL90 A0A1Q3FFI6 A0A1B6KDW2 A0A2M3ZYQ1 W5JK16 A0A2M3ZYG2 A0A2M4BLF1 A0A1B6IVK8 A0A0A9YM47 A0A1Y1KS02 A0A226ECT4 C0JBY4 R4WQK3 A0A0N8EAP0 A0A182GFX8 Q1HR78 B4L127 A0A3G5ART5 A0A3G5ANU4 B4PIY3 B4QKK6 A0A1W7R9Z2 A0A3B0J7U5 B4HI55 A0A1W4VP72 B3NHN4 T1L2F7 Q2M1A2 X2JGP4 A0A2I9LPF6 P54399 A0A2M3ZE69 B3M733 A0A1Q3FFB3 B4GUE5 E2BG03 B4N522 D3TRE3 B4LGU3 A0A1A9V3N0 B4IXK8 A0A0J7L6A5 A0A2Z5WPU1 A0A0P4VT63
EC Number
5.3.4.1
Pubmed
19121390
11892983
22651552
26354079
23622113
21967427
+ More
22118469 28756777 25244985 25348373 12364791 20966253 25315136 24495485 24438588 18362917 19820115 24845553 20920257 23761445 25401762 28004739 23691247 26483478 17204158 17510324 17994087 17550304 22936249 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 29248469 7787847 12537569 20798317 20353571 27129103
22118469 28756777 25244985 25348373 12364791 20966253 25315136 24495485 24438588 18362917 19820115 24845553 20920257 23761445 25401762 28004739 23691247 26483478 17204158 17510324 17994087 17550304 22936249 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 29248469 7787847 12537569 20798317 20353571 27129103
EMBL
BABH01012177
AF325211
AAG45936.1
AK401171
KQ459590
BAM17793.1
+ More
KPI97250.1 NWSH01002655 PCG67756.1 AK402305 BAM18927.1 JX183988 AGF69546.1 ODYU01004447 SOQ44326.1 GAIX01012014 JAA80546.1 JF777457 AEU11802.1 AGBW02003612 OWR55527.1 KZ149984 PZC75730.1 GALX01002703 JAB65763.1 CVRI01000042 CRK95503.1 GFDF01004054 JAV10030.1 GDRN01036013 JAI67374.1 GFDF01004053 JAV10031.1 GANO01001143 JAB58728.1 GFDF01004052 JAV10032.1 DS231920 EDS26398.1 GAKP01014585 JAC44367.1 DS232318 EDS39919.1 GDHF01033678 JAI18636.1 UFQT01000187 SSX21338.1 JN863961 AEW66914.1 AAAB01008986 EAL38666.3 APCN01003309 GDRN01036014 JAI67373.1 AXCM01006744 NEVH01007823 PNF35251.1 GFDG01001402 JAV17397.1 KA647381 AFP62010.1 GFDG01001401 JAV17398.1 GAMC01017431 GAMC01017428 JAB89124.1 AXCN02001026 GANO01000727 JAB59144.1 ATLV01023952 KE525347 KFB50164.1 GFDG01001400 JAV17399.1 KQ971321 EFA00647.1 KK852567 KDR21216.1 GAMD01002863 JAA98727.1 GGFK01000221 MBW33542.1 GGFJ01004696 MBW53837.1 GFDL01008773 JAV26272.1 GEBQ01030325 JAT09652.1 GGFK01000217 MBW33538.1 ADMH02001279 ETN63240.1 GGFK01000273 MBW33594.1 GGFJ01004734 MBW53875.1 GECU01021031 GECU01016836 GECU01003704 JAS86675.1 JAS90870.1 JAT04003.1 GBHO01009502 GBHO01009500 GBRD01012827 JAG34102.1 JAG34104.1 JAG52999.1 GEZM01077735 JAV63191.1 LNIX01000004 OXA55402.1 FJ179395 ACN89260.1 AK416967 BAN20182.1 GDIQ01045348 JAN49389.1 JXUM01060637 JXUM01060638 KQ562111 KXJ76669.1 DQ440216 CH477198 ABF18249.1 EAT48285.1 CH933809 EDW18184.1 MH979838 AYV89293.1 MH990459 AYV89006.1 CM000159 EDW94574.1 CM000363 CM002912 EDX10517.1 KMY99709.1 GFAH01000417 JAV47972.1 OUUW01000002 SPP76002.1 CH480815 EDW41551.1 CH954178 EDV51829.1 CAEY01000947 CH379069 EAL30673.1 AE014296 AHN58095.1 GFWZ01000275 MBW20265.1 U18973 BT001544 BT003181 BT011488 BT012439 GGFM01006048 MBW26799.1 CH902618 EDV40898.1 GFDL01008799 JAV26246.1 CH479191 EDW26228.1 GL448096 EFN85405.1 CH964101 EDW79461.1 CCAG010000249 EZ423995 ADD20271.1 CH940647 EDW70558.1 CH916366 EDV97470.1 LBMM01000553 KMQ98073.1 LC164244 BBC20867.1 GDKW01002455 JAI54140.1
KPI97250.1 NWSH01002655 PCG67756.1 AK402305 BAM18927.1 JX183988 AGF69546.1 ODYU01004447 SOQ44326.1 GAIX01012014 JAA80546.1 JF777457 AEU11802.1 AGBW02003612 OWR55527.1 KZ149984 PZC75730.1 GALX01002703 JAB65763.1 CVRI01000042 CRK95503.1 GFDF01004054 JAV10030.1 GDRN01036013 JAI67374.1 GFDF01004053 JAV10031.1 GANO01001143 JAB58728.1 GFDF01004052 JAV10032.1 DS231920 EDS26398.1 GAKP01014585 JAC44367.1 DS232318 EDS39919.1 GDHF01033678 JAI18636.1 UFQT01000187 SSX21338.1 JN863961 AEW66914.1 AAAB01008986 EAL38666.3 APCN01003309 GDRN01036014 JAI67373.1 AXCM01006744 NEVH01007823 PNF35251.1 GFDG01001402 JAV17397.1 KA647381 AFP62010.1 GFDG01001401 JAV17398.1 GAMC01017431 GAMC01017428 JAB89124.1 AXCN02001026 GANO01000727 JAB59144.1 ATLV01023952 KE525347 KFB50164.1 GFDG01001400 JAV17399.1 KQ971321 EFA00647.1 KK852567 KDR21216.1 GAMD01002863 JAA98727.1 GGFK01000221 MBW33542.1 GGFJ01004696 MBW53837.1 GFDL01008773 JAV26272.1 GEBQ01030325 JAT09652.1 GGFK01000217 MBW33538.1 ADMH02001279 ETN63240.1 GGFK01000273 MBW33594.1 GGFJ01004734 MBW53875.1 GECU01021031 GECU01016836 GECU01003704 JAS86675.1 JAS90870.1 JAT04003.1 GBHO01009502 GBHO01009500 GBRD01012827 JAG34102.1 JAG34104.1 JAG52999.1 GEZM01077735 JAV63191.1 LNIX01000004 OXA55402.1 FJ179395 ACN89260.1 AK416967 BAN20182.1 GDIQ01045348 JAN49389.1 JXUM01060637 JXUM01060638 KQ562111 KXJ76669.1 DQ440216 CH477198 ABF18249.1 EAT48285.1 CH933809 EDW18184.1 MH979838 AYV89293.1 MH990459 AYV89006.1 CM000159 EDW94574.1 CM000363 CM002912 EDX10517.1 KMY99709.1 GFAH01000417 JAV47972.1 OUUW01000002 SPP76002.1 CH480815 EDW41551.1 CH954178 EDV51829.1 CAEY01000947 CH379069 EAL30673.1 AE014296 AHN58095.1 GFWZ01000275 MBW20265.1 U18973 BT001544 BT003181 BT011488 BT012439 GGFM01006048 MBW26799.1 CH902618 EDV40898.1 GFDL01008799 JAV26246.1 CH479191 EDW26228.1 GL448096 EFN85405.1 CH964101 EDW79461.1 CCAG010000249 EZ423995 ADD20271.1 CH940647 EDW70558.1 CH916366 EDV97470.1 LBMM01000553 KMQ98073.1 LC164244 BBC20867.1 GDKW01002455 JAI54140.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000183832
UP000076408
+ More
UP000002320 UP000075884 UP000075902 UP000007062 UP000075882 UP000075840 UP000075900 UP000076407 UP000075903 UP000075885 UP000075883 UP000095301 UP000235965 UP000075920 UP000075886 UP000095300 UP000075880 UP000030765 UP000007266 UP000069272 UP000027135 UP000000673 UP000198287 UP000069940 UP000249989 UP000008820 UP000009192 UP000002282 UP000000304 UP000268350 UP000001292 UP000192221 UP000008711 UP000015104 UP000001819 UP000000803 UP000007801 UP000008744 UP000008237 UP000007798 UP000092444 UP000008792 UP000078200 UP000001070 UP000036403
UP000002320 UP000075884 UP000075902 UP000007062 UP000075882 UP000075840 UP000075900 UP000076407 UP000075903 UP000075885 UP000075883 UP000095301 UP000235965 UP000075920 UP000075886 UP000095300 UP000075880 UP000030765 UP000007266 UP000069272 UP000027135 UP000000673 UP000198287 UP000069940 UP000249989 UP000008820 UP000009192 UP000002282 UP000000304 UP000268350 UP000001292 UP000192221 UP000008711 UP000015104 UP000001819 UP000000803 UP000007801 UP000008744 UP000008237 UP000007798 UP000092444 UP000008792 UP000078200 UP000001070 UP000036403
Pfam
PF00085 Thioredoxin
Interpro
SUPFAM
SSF52833
SSF52833
ProteinModelPortal
H9JQN8
Q9GPH2
I4DIQ3
A0A2A4J7L8
I4DLY7
A0A0U1VTU3
+ More
A0A2H1VU96 S4NP33 G9D554 A0A212FP73 A0A2W1BSJ9 V5GZ61 A0A1J1I589 A0A1L8DUD1 A0A0P4WI78 A0A1L8DUB0 U5EV80 A0A1L8DUB3 A0A182Y1G6 B0WFQ4 A0A182MYS9 A0A034VQH7 B0X3M7 A0A0K8TWH4 A0A336M5N1 G9JKY3 A0A182TD69 Q5TMX9 A0A1S4HAB3 A0A182KS99 A0A182HQD4 A0A182RD13 A0A182WZT9 A0A182UP09 A0A0P4WLN5 A0A182P5V3 A0A182MG57 A0A1I8MXQ8 A0A2J7R328 A0A1L8EF87 T1PF58 A0A1L8EFE1 A0A182WEX4 W8BHI2 A0A182QYN8 U5EXV6 A0A1I8P5Q9 A0A182J8G7 A0A084WIX0 A0A1L8EFM8 D6WHD6 A0A182FBC4 A0A067RBI3 T1E8D6 A0A2M3ZYI7 A0A2M4BL90 A0A1Q3FFI6 A0A1B6KDW2 A0A2M3ZYQ1 W5JK16 A0A2M3ZYG2 A0A2M4BLF1 A0A1B6IVK8 A0A0A9YM47 A0A1Y1KS02 A0A226ECT4 C0JBY4 R4WQK3 A0A0N8EAP0 A0A182GFX8 Q1HR78 B4L127 A0A3G5ART5 A0A3G5ANU4 B4PIY3 B4QKK6 A0A1W7R9Z2 A0A3B0J7U5 B4HI55 A0A1W4VP72 B3NHN4 T1L2F7 Q2M1A2 X2JGP4 A0A2I9LPF6 P54399 A0A2M3ZE69 B3M733 A0A1Q3FFB3 B4GUE5 E2BG03 B4N522 D3TRE3 B4LGU3 A0A1A9V3N0 B4IXK8 A0A0J7L6A5 A0A2Z5WPU1 A0A0P4VT63
A0A2H1VU96 S4NP33 G9D554 A0A212FP73 A0A2W1BSJ9 V5GZ61 A0A1J1I589 A0A1L8DUD1 A0A0P4WI78 A0A1L8DUB0 U5EV80 A0A1L8DUB3 A0A182Y1G6 B0WFQ4 A0A182MYS9 A0A034VQH7 B0X3M7 A0A0K8TWH4 A0A336M5N1 G9JKY3 A0A182TD69 Q5TMX9 A0A1S4HAB3 A0A182KS99 A0A182HQD4 A0A182RD13 A0A182WZT9 A0A182UP09 A0A0P4WLN5 A0A182P5V3 A0A182MG57 A0A1I8MXQ8 A0A2J7R328 A0A1L8EF87 T1PF58 A0A1L8EFE1 A0A182WEX4 W8BHI2 A0A182QYN8 U5EXV6 A0A1I8P5Q9 A0A182J8G7 A0A084WIX0 A0A1L8EFM8 D6WHD6 A0A182FBC4 A0A067RBI3 T1E8D6 A0A2M3ZYI7 A0A2M4BL90 A0A1Q3FFI6 A0A1B6KDW2 A0A2M3ZYQ1 W5JK16 A0A2M3ZYG2 A0A2M4BLF1 A0A1B6IVK8 A0A0A9YM47 A0A1Y1KS02 A0A226ECT4 C0JBY4 R4WQK3 A0A0N8EAP0 A0A182GFX8 Q1HR78 B4L127 A0A3G5ART5 A0A3G5ANU4 B4PIY3 B4QKK6 A0A1W7R9Z2 A0A3B0J7U5 B4HI55 A0A1W4VP72 B3NHN4 T1L2F7 Q2M1A2 X2JGP4 A0A2I9LPF6 P54399 A0A2M3ZE69 B3M733 A0A1Q3FFB3 B4GUE5 E2BG03 B4N522 D3TRE3 B4LGU3 A0A1A9V3N0 B4IXK8 A0A0J7L6A5 A0A2Z5WPU1 A0A0P4VT63
PDB
4EL1
E-value=1.82176e-136,
Score=1246
Ontologies
GO
Topology
Subcellular location
Endoplasmic reticulum lumen
SignalP
Position: 1 - 17,
Likelihood: 0.999395
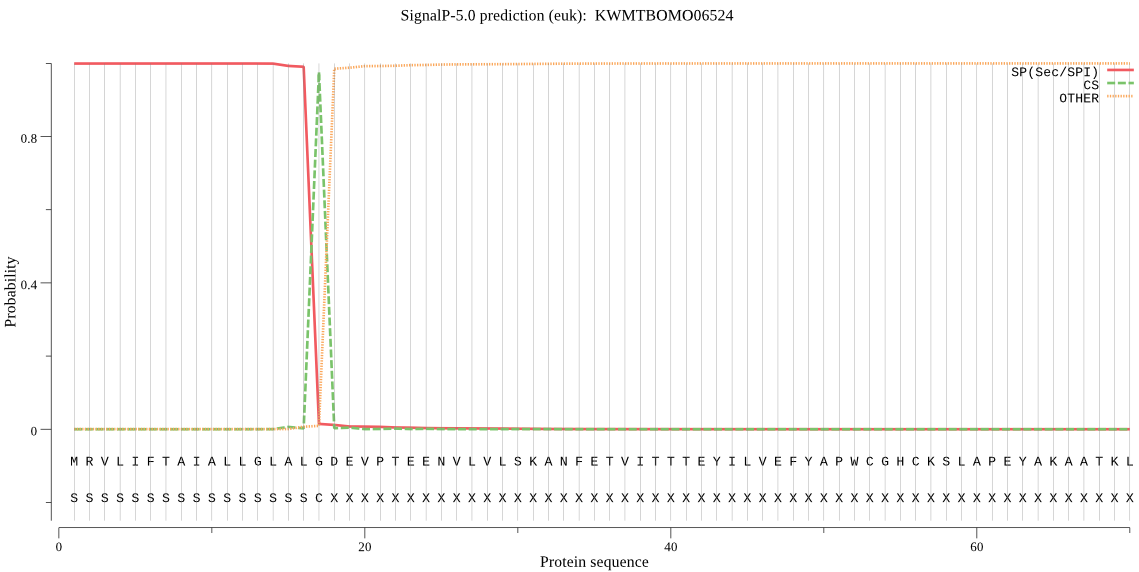
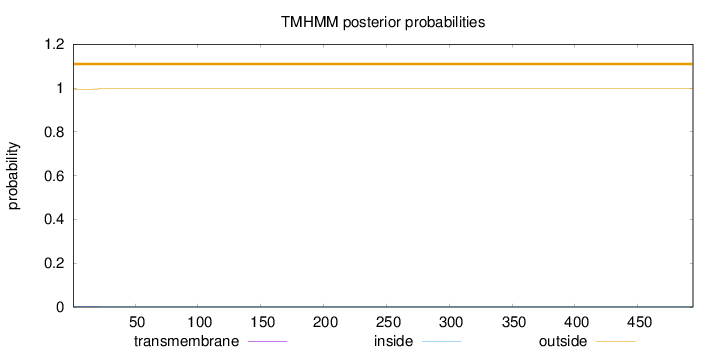
Length:
494
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07015
Exp number, first 60 AAs:
0.06996
Total prob of N-in:
0.00530
outside
1 - 494
Population Genetic Test Statistics
Pi
279.48187
Theta
190.171513
Tajima's D
2.14643
CLR
0.167676
CSRT
0.904904754762262
Interpretation
Uncertain
