Gene
KWMTBOMO06523 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011641
Annotation
PREDICTED:_target_of_Myb_protein_1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.327
Sequence
CDS
ATGTCGTTTTTTGGTGTAGGGAATCCCTTTTCTACGCCGGTGGGTCAAAAAATTGAACAAGCCACAGATGGCGCATTACCATCTGAAAATTGGGCTCTGAACATGGAAATATGCGATATAATAAACAGCAGTACGGACGGACCTAAAGATGCCATTAAAGCAATAAGAAAAAGGTTGACTACAAGCGCTGGGAAGAACTACACAGTGGTGATGTACACGCTGACTGTCCTGGAAACATGCGTAAAAAACTGTGGAAAGCCGTTCCATGTTCTCGTCTGCAATAAGGAATTTATATCAGAGTTGGTAAAATTGATTGGCCCCAAAAATGATCCGCCGACGGTGGTGCAAGACAAAGTATTGAGCCTCATCCAATGTTGGGCCGATGCCTTCCAGAACCAAGCTGAATTACAGGGAGTCGGTCAAGTGTACAACGAGCTTCGCACCAAAGGTGTTGAATTTCCGATGACCGACTTAGACGCCATGGCCCCGATTTTCACGCCACAGCGGAGTGTCCCGGATGGCGTTGAACAAGTTGGTTCCCCTCAGCGCTCCGTCTTGCCGCAGAACTCTCCTAGCAGGACAACTCCGGAACAGATCACTACTCAGGTGACGCTGAGCGACAGTCAGACCAACAAGCTGCGCGCCGACCTCGCCGTGGTGGAGGGGAACATGGCCGTCATGAACGACATGCTCAACGAGCTCACCGCCCGGCCGCAAGCAACGCGACACGACAGCGACGTCGAGCTTCTGAACGAGCTAGCGGACACGCTGCGTGCCATGCAGAGCCGCGTCGCCGAGCTCATAGCGCGCGTGGCGGCCGACGGCGCGCTCACCGCCGAGCTGCTGCACGCCAACGACCGCCTGCACAACCTGCTGCTGCGCCACGCGCGCTTCCTCAACAATAGGTCTGCAGCGGCGGCGGGCGCGGGGCCCACTCCGTCCGCCATCCTCGGCGCCGCCATGGGCGTGCCCGGCGCGGCGCCTACGTCACCAAAGAAACAAGAGGATGACGCCTTGATCGACCTAAGCGACGACGTCCCTGATATTGCGAAGCTGAGCGTAACGGATACTCAAGCAGAAAAGTCTCCGAGCAGCTCGAAGGATGAATTCGACATGTTCGCTCAATCGAGGAACATCACTTATGAAACCTCGAAGACAGGTACCAGCAGCTACGCCGATAACTTAGAAGACCAAACCACGACGAGCTTGGCCTCCGCCGCCAGCCACCGAGCCACACAGGGCCAACCTCTGGCGGCTTCCGAGAGCGTCCACGAGAGCGTGACCAGTAGCGACTTCGACAAGTTCCTGGCGGAGCGCGCGGCCGCCGCCGAGCAGCTGCCCGATATCAGCGACCGCAGCCCTAGGCACCGGCACATCAACAAGGATGACCAGGACTCTAGATATTGA
Protein
MSFFGVGNPFSTPVGQKIEQATDGALPSENWALNMEICDIINSSTDGPKDAIKAIRKRLTTSAGKNYTVVMYTLTVLETCVKNCGKPFHVLVCNKEFISELVKLIGPKNDPPTVVQDKVLSLIQCWADAFQNQAELQGVGQVYNELRTKGVEFPMTDLDAMAPIFTPQRSVPDGVEQVGSPQRSVLPQNSPSRTTPEQITTQVTLSDSQTNKLRADLAVVEGNMAVMNDMLNELTARPQATRHDSDVELLNELADTLRAMQSRVAELIARVAADGALTAELLHANDRLHNLLLRHARFLNNRSAAAAGAGPTPSAILGAAMGVPGAAPTSPKKQEDDALIDLSDDVPDIAKLSVTDTQAEKSPSSSKDEFDMFAQSRNITYETSKTGTSSYADNLEDQTTTSLASAASHRATQGQPLAASESVHESVTSSDFDKFLAERAAAAEQLPDISDRSPRHRHINKDDQDSRY
Summary
Uniprot
A0A2A4J6L0
A0A1E1WMT3
A0A1E1WNY6
A0A194PWW1
A0A212FP50
A0A2J7PJL2
+ More
D6WHD9 A0A067QWK9 A0A0L7QNF4 A0A1Y1MCE1 A0A2J7PJK9 A0A026WUC1 A0A1Y1MDN9 E9IVA6 E2AFD6 A0A3L8DR52 A0A1B6CYU7 A0A0J7KSH2 V9IG33 A0A310S4A2 A0A195DIW5 A0A0C9R7U5 E2B6W2 V5GT69 A0A087ZZE1 A0A195C0M1 A0A2A3EIP7 A0A195FHZ9 A0A151I115 A0A158NDC3 A0A0C9QTT2 A0A1B6F9U9 A0A1L8DW14 A0A224XNF5 A0A0V0G6E3 A0A023F392 A0A1L8DW12 A0A1L8DW00 U5EP22 A0A1B0CP56 A0A0P4VWL2 A0A069DUY2 Q17AJ9 A0A1S4FA26 A0A151XHJ3 A0A1Q3G3S4 A0A1Q3G3Y5 A0A1Q3G3R9 A0A1W7R9F1 A0A023ETE4 A0A1Q3G3U5 A0A1Q3G3S9 A0A1Q3G3S2 A0A154PID0 A0A1Q3G3S5 T1J1S5 T1HFP0 A0A0A9Z597 A0A0K8SEY3 A0A0K8SGD7 A0A1I8N323 A0A3Q0J0R2 A0A1W4WMT6 A0A1W4WBS9 A0A3Q0J5R5 A0A0M9A505 A0A1W4WBS5 A0A1W4WLX4 A0A087UT54 A0A182Q697 A0A1I8N338 A0A034VP97 W8C803 J9JLH3 A0A1Q3FGB6 A0A034VR48 A0A182Y352 A0A0P4WF80 Q7PNZ6 A0A2S2QI93 A0A182UUD2 A0A182KP48 K7ISH2 A0A2H8U0B3 A0A1B6LB82 A0A0A1XCG4 A0A182G7L1 A0A232FME8 A0A3F2Z155 A0A1A9W7L4 V5HHR2 A0A1W4U9B0 Q7PIF9 A0A1B0A353 A0A336M8V8 A0A1B6KQM5 A0A293N0H0 A0A1B0FGL1 D3TND3
D6WHD9 A0A067QWK9 A0A0L7QNF4 A0A1Y1MCE1 A0A2J7PJK9 A0A026WUC1 A0A1Y1MDN9 E9IVA6 E2AFD6 A0A3L8DR52 A0A1B6CYU7 A0A0J7KSH2 V9IG33 A0A310S4A2 A0A195DIW5 A0A0C9R7U5 E2B6W2 V5GT69 A0A087ZZE1 A0A195C0M1 A0A2A3EIP7 A0A195FHZ9 A0A151I115 A0A158NDC3 A0A0C9QTT2 A0A1B6F9U9 A0A1L8DW14 A0A224XNF5 A0A0V0G6E3 A0A023F392 A0A1L8DW12 A0A1L8DW00 U5EP22 A0A1B0CP56 A0A0P4VWL2 A0A069DUY2 Q17AJ9 A0A1S4FA26 A0A151XHJ3 A0A1Q3G3S4 A0A1Q3G3Y5 A0A1Q3G3R9 A0A1W7R9F1 A0A023ETE4 A0A1Q3G3U5 A0A1Q3G3S9 A0A1Q3G3S2 A0A154PID0 A0A1Q3G3S5 T1J1S5 T1HFP0 A0A0A9Z597 A0A0K8SEY3 A0A0K8SGD7 A0A1I8N323 A0A3Q0J0R2 A0A1W4WMT6 A0A1W4WBS9 A0A3Q0J5R5 A0A0M9A505 A0A1W4WBS5 A0A1W4WLX4 A0A087UT54 A0A182Q697 A0A1I8N338 A0A034VP97 W8C803 J9JLH3 A0A1Q3FGB6 A0A034VR48 A0A182Y352 A0A0P4WF80 Q7PNZ6 A0A2S2QI93 A0A182UUD2 A0A182KP48 K7ISH2 A0A2H8U0B3 A0A1B6LB82 A0A0A1XCG4 A0A182G7L1 A0A232FME8 A0A3F2Z155 A0A1A9W7L4 V5HHR2 A0A1W4U9B0 Q7PIF9 A0A1B0A353 A0A336M8V8 A0A1B6KQM5 A0A293N0H0 A0A1B0FGL1 D3TND3
Pubmed
EMBL
NWSH01002655
PCG67757.1
GDQN01002898
JAT88156.1
GDQN01002349
JAT88705.1
+ More
KQ459590 KPI97249.1 AGBW02003612 OWR55526.1 NEVH01024946 PNF16527.1 KQ971321 EFA00649.1 KK853282 KDR09020.1 KQ414855 KOC60148.1 GEZM01037733 JAV81965.1 PNF16526.1 KK107102 EZA59630.1 GEZM01037732 JAV81966.1 GL766194 EFZ15482.1 GL439108 EFN67829.1 QOIP01000005 RLU22871.1 GEDC01018649 JAS18649.1 LBMM01003750 KMQ93179.1 JR045756 AEY60048.1 KQ779356 OAD51972.1 KQ980800 KYN12820.1 GBYB01004100 JAG73867.1 GL446031 EFN88567.1 GALX01003624 JAB64842.1 KQ978379 KYM94397.1 KZ288230 PBC31597.1 KQ981560 KYN39981.1 KQ976599 KYM79504.1 ADTU01012589 ADTU01012590 GBYB01004097 JAG73864.1 GECZ01022760 GECZ01018546 JAS47009.1 JAS51223.1 GFDF01003468 JAV10616.1 GFTR01006743 JAW09683.1 GECL01003181 JAP02943.1 GBBI01003056 JAC15656.1 GFDF01003466 JAV10618.1 GFDF01003467 JAV10617.1 GANO01000348 JAB59523.1 AJWK01021441 GDKW01000080 JAI56515.1 GBGD01001253 JAC87636.1 CH477333 EAT43294.1 KQ982130 KYQ59827.1 GFDL01000590 JAV34455.1 GFDL01000569 JAV34476.1 GFDL01000592 JAV34453.1 GFAH01000629 JAV47760.1 GAPW01001035 JAC12563.1 GFDL01000567 JAV34478.1 GFDL01000591 JAV34454.1 GFDL01000593 JAV34452.1 KQ434924 KZC11615.1 GFDL01000595 JAV34450.1 JH431789 ACPB03012545 GBHO01038030 GBHO01004005 GDHC01008661 GDHC01005512 JAG05574.1 JAG39599.1 JAQ09968.1 JAQ13117.1 GBRD01013979 JAG51847.1 GBRD01013981 JAG51845.1 KQ435732 KOX77360.1 KK121471 KFM80543.1 AXCN02000943 GAKP01014678 JAC44274.1 GAMC01007741 JAB98814.1 ABLF02039935 GFDL01008503 JAV26542.1 GAKP01014677 JAC44275.1 GDRN01061212 GDRN01061211 JAI65231.1 AAAB01008960 EAA11281.5 GGMS01008266 MBY77469.1 GFXV01007646 MBW19451.1 GEBQ01019010 JAT20967.1 GBXI01011223 GBXI01006139 JAD03069.1 JAD08153.1 JXUM01150486 JXUM01150487 KQ571012 KXJ68261.1 NNAY01000041 OXU31628.1 GANP01001258 JAB83210.1 EAA44160.4 UFQS01000501 UFQT01000501 SSX04433.1 SSX24797.1 GEBQ01026234 JAT13743.1 GFWV01020167 MAA44895.1 CCAG010006947 EZ422935 ADD19211.1
KQ459590 KPI97249.1 AGBW02003612 OWR55526.1 NEVH01024946 PNF16527.1 KQ971321 EFA00649.1 KK853282 KDR09020.1 KQ414855 KOC60148.1 GEZM01037733 JAV81965.1 PNF16526.1 KK107102 EZA59630.1 GEZM01037732 JAV81966.1 GL766194 EFZ15482.1 GL439108 EFN67829.1 QOIP01000005 RLU22871.1 GEDC01018649 JAS18649.1 LBMM01003750 KMQ93179.1 JR045756 AEY60048.1 KQ779356 OAD51972.1 KQ980800 KYN12820.1 GBYB01004100 JAG73867.1 GL446031 EFN88567.1 GALX01003624 JAB64842.1 KQ978379 KYM94397.1 KZ288230 PBC31597.1 KQ981560 KYN39981.1 KQ976599 KYM79504.1 ADTU01012589 ADTU01012590 GBYB01004097 JAG73864.1 GECZ01022760 GECZ01018546 JAS47009.1 JAS51223.1 GFDF01003468 JAV10616.1 GFTR01006743 JAW09683.1 GECL01003181 JAP02943.1 GBBI01003056 JAC15656.1 GFDF01003466 JAV10618.1 GFDF01003467 JAV10617.1 GANO01000348 JAB59523.1 AJWK01021441 GDKW01000080 JAI56515.1 GBGD01001253 JAC87636.1 CH477333 EAT43294.1 KQ982130 KYQ59827.1 GFDL01000590 JAV34455.1 GFDL01000569 JAV34476.1 GFDL01000592 JAV34453.1 GFAH01000629 JAV47760.1 GAPW01001035 JAC12563.1 GFDL01000567 JAV34478.1 GFDL01000591 JAV34454.1 GFDL01000593 JAV34452.1 KQ434924 KZC11615.1 GFDL01000595 JAV34450.1 JH431789 ACPB03012545 GBHO01038030 GBHO01004005 GDHC01008661 GDHC01005512 JAG05574.1 JAG39599.1 JAQ09968.1 JAQ13117.1 GBRD01013979 JAG51847.1 GBRD01013981 JAG51845.1 KQ435732 KOX77360.1 KK121471 KFM80543.1 AXCN02000943 GAKP01014678 JAC44274.1 GAMC01007741 JAB98814.1 ABLF02039935 GFDL01008503 JAV26542.1 GAKP01014677 JAC44275.1 GDRN01061212 GDRN01061211 JAI65231.1 AAAB01008960 EAA11281.5 GGMS01008266 MBY77469.1 GFXV01007646 MBW19451.1 GEBQ01019010 JAT20967.1 GBXI01011223 GBXI01006139 JAD03069.1 JAD08153.1 JXUM01150486 JXUM01150487 KQ571012 KXJ68261.1 NNAY01000041 OXU31628.1 GANP01001258 JAB83210.1 EAA44160.4 UFQS01000501 UFQT01000501 SSX04433.1 SSX24797.1 GEBQ01026234 JAT13743.1 GFWV01020167 MAA44895.1 CCAG010006947 EZ422935 ADD19211.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000235965
UP000007266
UP000027135
+ More
UP000053825 UP000053097 UP000000311 UP000279307 UP000036403 UP000078492 UP000008237 UP000005203 UP000078542 UP000242457 UP000078541 UP000078540 UP000005205 UP000092461 UP000008820 UP000075809 UP000076502 UP000015103 UP000095301 UP000079169 UP000192223 UP000053105 UP000054359 UP000075886 UP000007819 UP000076408 UP000007062 UP000075903 UP000075882 UP000002358 UP000069940 UP000249989 UP000215335 UP000075920 UP000091820 UP000192221 UP000092445 UP000092444
UP000053825 UP000053097 UP000000311 UP000279307 UP000036403 UP000078492 UP000008237 UP000005203 UP000078542 UP000242457 UP000078541 UP000078540 UP000005205 UP000092461 UP000008820 UP000075809 UP000076502 UP000015103 UP000095301 UP000079169 UP000192223 UP000053105 UP000054359 UP000075886 UP000007819 UP000076408 UP000007062 UP000075903 UP000075882 UP000002358 UP000069940 UP000249989 UP000215335 UP000075920 UP000091820 UP000192221 UP000092445 UP000092444
Interpro
SUPFAM
SSF48464
SSF48464
Gene 3D
ProteinModelPortal
A0A2A4J6L0
A0A1E1WMT3
A0A1E1WNY6
A0A194PWW1
A0A212FP50
A0A2J7PJL2
+ More
D6WHD9 A0A067QWK9 A0A0L7QNF4 A0A1Y1MCE1 A0A2J7PJK9 A0A026WUC1 A0A1Y1MDN9 E9IVA6 E2AFD6 A0A3L8DR52 A0A1B6CYU7 A0A0J7KSH2 V9IG33 A0A310S4A2 A0A195DIW5 A0A0C9R7U5 E2B6W2 V5GT69 A0A087ZZE1 A0A195C0M1 A0A2A3EIP7 A0A195FHZ9 A0A151I115 A0A158NDC3 A0A0C9QTT2 A0A1B6F9U9 A0A1L8DW14 A0A224XNF5 A0A0V0G6E3 A0A023F392 A0A1L8DW12 A0A1L8DW00 U5EP22 A0A1B0CP56 A0A0P4VWL2 A0A069DUY2 Q17AJ9 A0A1S4FA26 A0A151XHJ3 A0A1Q3G3S4 A0A1Q3G3Y5 A0A1Q3G3R9 A0A1W7R9F1 A0A023ETE4 A0A1Q3G3U5 A0A1Q3G3S9 A0A1Q3G3S2 A0A154PID0 A0A1Q3G3S5 T1J1S5 T1HFP0 A0A0A9Z597 A0A0K8SEY3 A0A0K8SGD7 A0A1I8N323 A0A3Q0J0R2 A0A1W4WMT6 A0A1W4WBS9 A0A3Q0J5R5 A0A0M9A505 A0A1W4WBS5 A0A1W4WLX4 A0A087UT54 A0A182Q697 A0A1I8N338 A0A034VP97 W8C803 J9JLH3 A0A1Q3FGB6 A0A034VR48 A0A182Y352 A0A0P4WF80 Q7PNZ6 A0A2S2QI93 A0A182UUD2 A0A182KP48 K7ISH2 A0A2H8U0B3 A0A1B6LB82 A0A0A1XCG4 A0A182G7L1 A0A232FME8 A0A3F2Z155 A0A1A9W7L4 V5HHR2 A0A1W4U9B0 Q7PIF9 A0A1B0A353 A0A336M8V8 A0A1B6KQM5 A0A293N0H0 A0A1B0FGL1 D3TND3
D6WHD9 A0A067QWK9 A0A0L7QNF4 A0A1Y1MCE1 A0A2J7PJK9 A0A026WUC1 A0A1Y1MDN9 E9IVA6 E2AFD6 A0A3L8DR52 A0A1B6CYU7 A0A0J7KSH2 V9IG33 A0A310S4A2 A0A195DIW5 A0A0C9R7U5 E2B6W2 V5GT69 A0A087ZZE1 A0A195C0M1 A0A2A3EIP7 A0A195FHZ9 A0A151I115 A0A158NDC3 A0A0C9QTT2 A0A1B6F9U9 A0A1L8DW14 A0A224XNF5 A0A0V0G6E3 A0A023F392 A0A1L8DW12 A0A1L8DW00 U5EP22 A0A1B0CP56 A0A0P4VWL2 A0A069DUY2 Q17AJ9 A0A1S4FA26 A0A151XHJ3 A0A1Q3G3S4 A0A1Q3G3Y5 A0A1Q3G3R9 A0A1W7R9F1 A0A023ETE4 A0A1Q3G3U5 A0A1Q3G3S9 A0A1Q3G3S2 A0A154PID0 A0A1Q3G3S5 T1J1S5 T1HFP0 A0A0A9Z597 A0A0K8SEY3 A0A0K8SGD7 A0A1I8N323 A0A3Q0J0R2 A0A1W4WMT6 A0A1W4WBS9 A0A3Q0J5R5 A0A0M9A505 A0A1W4WBS5 A0A1W4WLX4 A0A087UT54 A0A182Q697 A0A1I8N338 A0A034VP97 W8C803 J9JLH3 A0A1Q3FGB6 A0A034VR48 A0A182Y352 A0A0P4WF80 Q7PNZ6 A0A2S2QI93 A0A182UUD2 A0A182KP48 K7ISH2 A0A2H8U0B3 A0A1B6LB82 A0A0A1XCG4 A0A182G7L1 A0A232FME8 A0A3F2Z155 A0A1A9W7L4 V5HHR2 A0A1W4U9B0 Q7PIF9 A0A1B0A353 A0A336M8V8 A0A1B6KQM5 A0A293N0H0 A0A1B0FGL1 D3TND3
PDB
1ELK
E-value=8.07614e-52,
Score=516
Ontologies
GO
Topology
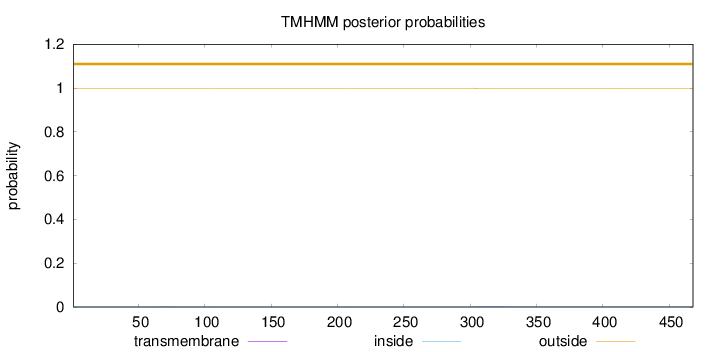
Length:
468
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04222
Exp number, first 60 AAs:
0.0001
Total prob of N-in:
0.00050
outside
1 - 468
Population Genetic Test Statistics
Pi
200.35975
Theta
160.047951
Tajima's D
0.739947
CLR
0.374445
CSRT
0.584320783960802
Interpretation
Uncertain
