Gene
KWMTBOMO06521 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011639
Annotation
enoyl-CoA_delta_isomerase_1_[Agrotis_segetum]
Location in the cell
Mitochondrial Reliability : 1.995
Sequence
CDS
ATGTTTCCTCTTCGCCAAGTAATTGCAACTGTACGCTATACAACTCCAGCATTCCGATCAATGTCAGCAAGTACAGGTCCTTTAATAGACTTGGCAGTTGATAATGACGGTATAGCAACATTAACCATGCAGAGACCTCCGGTAAACAGTCTGAACTTGGAATTGTTAGATGCATTAGGCAAAGCACTTGATGATGTATCTAAAAACAAGTCCAGGGGCGTCGTCTTGACATCGTCATCACCTACAGTATTCTCTGCTGGTCTGGACATTATGGAGATGTACAAACCTGATATAAAAAGAGTTGAAACATTCTGGACAACATTACAAGAAGTCTGGCTTAAATTATTTGGATCCAACTACCCTACTGCTGCTGCAATTAATGGTCATGCACCTGCTGGTGGATGTCTCCTGTCAATGTCCTGTGAATACAGAGCTATGGTCACAGGGAAATACAACATTGGTTTGAATGAAACAGCATTGGGCATTGTTGCTCCAAAGTGGTTTATGGACACTATGTGTAATACTATATCTAAGAGAGAGGCTGAATATGCACTCACCACAGCTAGAATGTTCAACACTGATGAAGCATTGAAGGTGGGACTCATCGACGTTTTAGCTACAGACAAGGCTGATGCGGTTGATAAATGCAAGAAGTTTATAAAGAAATTCGACAAAATCTCGCCTATAGCTCGTGCTTTAACCAAGCAAAAGATCAGGGCAGAACCTTTGACGTGGCTCATCAAGAATCGTAAACAGGACACTGATGAGTTTTTAACTTTTGTACAGAACCCGAAAGTACAACAATCGTTAGAACTTTACATTCAAGCTCTCAAACAGAAATCTGCTAAATAA
Protein
MFPLRQVIATVRYTTPAFRSMSASTGPLIDLAVDNDGIATLTMQRPPVNSLNLELLDALGKALDDVSKNKSRGVVLTSSSPTVFSAGLDIMEMYKPDIKRVETFWTTLQEVWLKLFGSNYPTAAAINGHAPAGGCLLSMSCEYRAMVTGKYNIGLNETALGIVAPKWFMDTMCNTISKREAEYALTTARMFNTDEALKVGLIDVLATDKADAVDKCKKFIKKFDKISPIARALTKQKIRAEPLTWLIKNRKQDTDEFLTFVQNPKVQQSLELYIQALKQKSAK
Summary
Similarity
Belongs to the enoyl-CoA hydratase/isomerase family.
Uniprot
A0A2A4J8K4
A0A2A4J706
A0A2W1BQ86
A0A068FMQ6
D2XML5
H9JQ33
+ More
A0A2H1W6W5 A0A194PV20 A0A0N1IDE6 A0A212FP47 I4DLD0 A0A067QJN4 A0A1Q3F4E6 A0A1S4F8J4 Q17BZ3 B0X2M5 A0A1B6EQM4 A0A182M893 A0A023EME0 A0A182WXJ1 A0A182GKN4 A0A182Q126 A0A182HQX3 Q5TNW4 A0A182NNP0 A0A182S7A2 A0A182W8E3 A0A182YQV3 A0A1W4WA15 B3N8K0 A0A182PTL4 A0A2M4AER7 B4NYP0 B4MZ14 A0A2M3Z879 A0A1L8EI30 A0A2M4BW00 T1E8C9 A0A336MF19 A0A084VWW1 A0A182FN10 A0A1L8EIB6 B4JE40 B4G7V5 A0A2M3ZI06 B3MPA8 A0A1Y1K631 A0A3B0JU57 Q9VL68 A0A1A9ZVU0 A0A0M3QT57 Q29NE3 A0A0L0CHR4 B4HWC5 A0A1A9V2B2 W5JM77 B4Q896 A0A1A9XJD9 A0A1I8P1X3 A0A1B0BLS1 V5GVQ4 A0A1I8N871 A0A1I8N874 A0A1B6CS95 B4LRD3 U5ESR8 D6WHD7 A0A1W4WXT2 D3TPU1 B4KJE7 A0A0A9WCF2 A0A1B0DDP7 A0A0N7Z960 R4G430 A0A023F9D3 A0A232FMN9 A0A1L8DZ86 A0A0V0G5L7 A0A1A9W5B1 B5DJ49 A0A224XHI7 A0A182R1M1 A0A0A1WIN6 J3JU08 A0A069DQU1 D1FPK3 A0A3B0JX44 A0A2R7VRE1 J9K4B4 A0A182NNN9 W8CCQ1 A0A0C9QFR4 A0A034VW47 A0A026WX28 A0A1B6L2U0 A0A0K8TWG5 A0A0L7QNE7 A0A195BY33 B4NYP1
A0A2H1W6W5 A0A194PV20 A0A0N1IDE6 A0A212FP47 I4DLD0 A0A067QJN4 A0A1Q3F4E6 A0A1S4F8J4 Q17BZ3 B0X2M5 A0A1B6EQM4 A0A182M893 A0A023EME0 A0A182WXJ1 A0A182GKN4 A0A182Q126 A0A182HQX3 Q5TNW4 A0A182NNP0 A0A182S7A2 A0A182W8E3 A0A182YQV3 A0A1W4WA15 B3N8K0 A0A182PTL4 A0A2M4AER7 B4NYP0 B4MZ14 A0A2M3Z879 A0A1L8EI30 A0A2M4BW00 T1E8C9 A0A336MF19 A0A084VWW1 A0A182FN10 A0A1L8EIB6 B4JE40 B4G7V5 A0A2M3ZI06 B3MPA8 A0A1Y1K631 A0A3B0JU57 Q9VL68 A0A1A9ZVU0 A0A0M3QT57 Q29NE3 A0A0L0CHR4 B4HWC5 A0A1A9V2B2 W5JM77 B4Q896 A0A1A9XJD9 A0A1I8P1X3 A0A1B0BLS1 V5GVQ4 A0A1I8N871 A0A1I8N874 A0A1B6CS95 B4LRD3 U5ESR8 D6WHD7 A0A1W4WXT2 D3TPU1 B4KJE7 A0A0A9WCF2 A0A1B0DDP7 A0A0N7Z960 R4G430 A0A023F9D3 A0A232FMN9 A0A1L8DZ86 A0A0V0G5L7 A0A1A9W5B1 B5DJ49 A0A224XHI7 A0A182R1M1 A0A0A1WIN6 J3JU08 A0A069DQU1 D1FPK3 A0A3B0JX44 A0A2R7VRE1 J9K4B4 A0A182NNN9 W8CCQ1 A0A0C9QFR4 A0A034VW47 A0A026WX28 A0A1B6L2U0 A0A0K8TWG5 A0A0L7QNE7 A0A195BY33 B4NYP1
Pubmed
28756777
26385554
20074338
19121390
26354079
22118469
+ More
22651552 24845553 17510324 24945155 26483478 12364791 14747013 17210077 25244985 17994087 17550304 24438588 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23185243 26108605 20920257 23761445 22936249 25315136 18362917 19820115 20353571 25401762 26823975 27129103 25474469 28648823 25830018 22516182 23537049 26334808 20441151 24495485 25348373 24508170 30249741
22651552 24845553 17510324 24945155 26483478 12364791 14747013 17210077 25244985 17994087 17550304 24438588 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23185243 26108605 20920257 23761445 22936249 25315136 18362917 19820115 20353571 25401762 26823975 27129103 25474469 28648823 25830018 22516182 23537049 26334808 20441151 24495485 25348373 24508170 30249741
EMBL
NWSH01002655
PCG67742.1
PCG67741.1
KZ149984
PZC75735.1
KJ622111
+ More
AID66701.1 GU205161 ADB57048.1 BABH01012182 ODYU01006705 SOQ48787.1 KQ459590 KPI97246.1 KQ461161 KPJ08044.1 AGBW02003612 OWR55524.1 AK402098 BAM18720.1 KK853282 KDR09023.1 GFDL01012610 JAV22435.1 CH477314 EAT43826.1 DS232291 EDS39248.1 GECZ01029518 GECZ01025086 GECZ01021425 GECZ01007805 GECZ01007455 GECZ01007410 JAS40251.1 JAS44683.1 JAS48344.1 JAS61964.1 JAS62314.1 JAS62359.1 AXCM01015334 GAPW01003121 JAC10477.1 JXUM01070277 KQ562597 KXJ75516.1 AXCN02001968 APCN01001609 AAAB01008980 EAL39276.3 CH954177 EDV58423.1 GGFK01005807 MBW39128.1 CM000157 EDW88704.1 KRJ97985.1 CH963913 EDW77410.1 GGFM01003954 MBW24705.1 GFDG01000432 JAV18367.1 GGFJ01008119 MBW57260.1 GAMD01002287 JAA99303.1 UFQT01000822 SSX27393.1 ATLV01017759 KE525185 KFB42455.1 GFDG01000433 JAV18366.1 CH916368 EDW03560.1 CH479180 EDW28453.1 GGFM01007402 MBW28153.1 CH902620 EDV32227.1 GEZM01091399 JAV56854.1 OUUW01000010 SPP85644.1 AE014134 AY113635 BT015179 AAF52828.2 AAM29640.1 AAT94408.1 AGB92839.1 CP012523 ALC38333.1 CH379060 EAL33399.1 KRT03795.1 JRES01000384 KNC31757.1 CH480818 EDW52320.1 ADMH02000643 ETN65467.1 CM000361 CM002910 EDX04418.1 KMY89359.1 JXJN01016540 GALX01004143 JAB64323.1 GEDC01020889 JAS16409.1 CH940649 EDW64603.1 GANO01003110 JAB56761.1 KQ971321 EFA00648.1 CCAG010013819 EZ423443 ADD19719.1 CH933807 EDW12522.1 GBHO01039416 GBRD01001755 GBRD01001754 GDHC01007805 JAG04188.1 JAG64066.1 JAQ10824.1 AJVK01032138 GDKW01001451 JAI55144.1 ACPB03012545 GAHY01001146 JAA76364.1 GBBI01001189 JAC17523.1 NNAY01000041 OXU31627.1 GFDF01002489 JAV11595.1 GECL01002822 JAP03302.1 EDY69918.1 KRT03794.1 GFTR01004470 JAW11956.1 GBXI01015751 JAC98540.1 BT126719 KB631539 KB632107 AEE61681.1 ERL84357.1 ERL88881.1 GBGD01002471 JAC86418.1 EZ419753 ACY69926.1 SPP85643.1 KK854036 PTY09976.1 ABLF02019997 GAMC01003306 JAC03250.1 GBYB01013333 JAG83100.1 GAKP01012877 JAC46075.1 KK107078 QOIP01000011 EZA60296.1 RLU16951.1 GEBQ01021925 JAT18052.1 GDHF01033527 JAI18787.1 KQ414855 KOC60150.1 KQ978501 KYM93502.1 EDW88705.1
AID66701.1 GU205161 ADB57048.1 BABH01012182 ODYU01006705 SOQ48787.1 KQ459590 KPI97246.1 KQ461161 KPJ08044.1 AGBW02003612 OWR55524.1 AK402098 BAM18720.1 KK853282 KDR09023.1 GFDL01012610 JAV22435.1 CH477314 EAT43826.1 DS232291 EDS39248.1 GECZ01029518 GECZ01025086 GECZ01021425 GECZ01007805 GECZ01007455 GECZ01007410 JAS40251.1 JAS44683.1 JAS48344.1 JAS61964.1 JAS62314.1 JAS62359.1 AXCM01015334 GAPW01003121 JAC10477.1 JXUM01070277 KQ562597 KXJ75516.1 AXCN02001968 APCN01001609 AAAB01008980 EAL39276.3 CH954177 EDV58423.1 GGFK01005807 MBW39128.1 CM000157 EDW88704.1 KRJ97985.1 CH963913 EDW77410.1 GGFM01003954 MBW24705.1 GFDG01000432 JAV18367.1 GGFJ01008119 MBW57260.1 GAMD01002287 JAA99303.1 UFQT01000822 SSX27393.1 ATLV01017759 KE525185 KFB42455.1 GFDG01000433 JAV18366.1 CH916368 EDW03560.1 CH479180 EDW28453.1 GGFM01007402 MBW28153.1 CH902620 EDV32227.1 GEZM01091399 JAV56854.1 OUUW01000010 SPP85644.1 AE014134 AY113635 BT015179 AAF52828.2 AAM29640.1 AAT94408.1 AGB92839.1 CP012523 ALC38333.1 CH379060 EAL33399.1 KRT03795.1 JRES01000384 KNC31757.1 CH480818 EDW52320.1 ADMH02000643 ETN65467.1 CM000361 CM002910 EDX04418.1 KMY89359.1 JXJN01016540 GALX01004143 JAB64323.1 GEDC01020889 JAS16409.1 CH940649 EDW64603.1 GANO01003110 JAB56761.1 KQ971321 EFA00648.1 CCAG010013819 EZ423443 ADD19719.1 CH933807 EDW12522.1 GBHO01039416 GBRD01001755 GBRD01001754 GDHC01007805 JAG04188.1 JAG64066.1 JAQ10824.1 AJVK01032138 GDKW01001451 JAI55144.1 ACPB03012545 GAHY01001146 JAA76364.1 GBBI01001189 JAC17523.1 NNAY01000041 OXU31627.1 GFDF01002489 JAV11595.1 GECL01002822 JAP03302.1 EDY69918.1 KRT03794.1 GFTR01004470 JAW11956.1 GBXI01015751 JAC98540.1 BT126719 KB631539 KB632107 AEE61681.1 ERL84357.1 ERL88881.1 GBGD01002471 JAC86418.1 EZ419753 ACY69926.1 SPP85643.1 KK854036 PTY09976.1 ABLF02019997 GAMC01003306 JAC03250.1 GBYB01013333 JAG83100.1 GAKP01012877 JAC46075.1 KK107078 QOIP01000011 EZA60296.1 RLU16951.1 GEBQ01021925 JAT18052.1 GDHF01033527 JAI18787.1 KQ414855 KOC60150.1 KQ978501 KYM93502.1 EDW88705.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000053240
UP000007151
UP000027135
+ More
UP000008820 UP000002320 UP000075883 UP000076407 UP000069940 UP000249989 UP000075886 UP000075840 UP000007062 UP000075884 UP000075901 UP000075920 UP000076408 UP000192221 UP000008711 UP000075885 UP000002282 UP000007798 UP000030765 UP000069272 UP000001070 UP000008744 UP000007801 UP000268350 UP000000803 UP000092445 UP000092553 UP000001819 UP000037069 UP000001292 UP000078200 UP000000673 UP000000304 UP000092443 UP000095300 UP000092460 UP000095301 UP000008792 UP000007266 UP000192223 UP000092444 UP000009192 UP000092462 UP000015103 UP000215335 UP000091820 UP000075900 UP000030742 UP000007819 UP000053097 UP000279307 UP000053825 UP000078542
UP000008820 UP000002320 UP000075883 UP000076407 UP000069940 UP000249989 UP000075886 UP000075840 UP000007062 UP000075884 UP000075901 UP000075920 UP000076408 UP000192221 UP000008711 UP000075885 UP000002282 UP000007798 UP000030765 UP000069272 UP000001070 UP000008744 UP000007801 UP000268350 UP000000803 UP000092445 UP000092553 UP000001819 UP000037069 UP000001292 UP000078200 UP000000673 UP000000304 UP000092443 UP000095300 UP000092460 UP000095301 UP000008792 UP000007266 UP000192223 UP000092444 UP000009192 UP000092462 UP000015103 UP000215335 UP000091820 UP000075900 UP000030742 UP000007819 UP000053097 UP000279307 UP000053825 UP000078542
Pfam
PF00378 ECH_1
Interpro
SUPFAM
SSF52096
SSF52096
Gene 3D
ProteinModelPortal
A0A2A4J8K4
A0A2A4J706
A0A2W1BQ86
A0A068FMQ6
D2XML5
H9JQ33
+ More
A0A2H1W6W5 A0A194PV20 A0A0N1IDE6 A0A212FP47 I4DLD0 A0A067QJN4 A0A1Q3F4E6 A0A1S4F8J4 Q17BZ3 B0X2M5 A0A1B6EQM4 A0A182M893 A0A023EME0 A0A182WXJ1 A0A182GKN4 A0A182Q126 A0A182HQX3 Q5TNW4 A0A182NNP0 A0A182S7A2 A0A182W8E3 A0A182YQV3 A0A1W4WA15 B3N8K0 A0A182PTL4 A0A2M4AER7 B4NYP0 B4MZ14 A0A2M3Z879 A0A1L8EI30 A0A2M4BW00 T1E8C9 A0A336MF19 A0A084VWW1 A0A182FN10 A0A1L8EIB6 B4JE40 B4G7V5 A0A2M3ZI06 B3MPA8 A0A1Y1K631 A0A3B0JU57 Q9VL68 A0A1A9ZVU0 A0A0M3QT57 Q29NE3 A0A0L0CHR4 B4HWC5 A0A1A9V2B2 W5JM77 B4Q896 A0A1A9XJD9 A0A1I8P1X3 A0A1B0BLS1 V5GVQ4 A0A1I8N871 A0A1I8N874 A0A1B6CS95 B4LRD3 U5ESR8 D6WHD7 A0A1W4WXT2 D3TPU1 B4KJE7 A0A0A9WCF2 A0A1B0DDP7 A0A0N7Z960 R4G430 A0A023F9D3 A0A232FMN9 A0A1L8DZ86 A0A0V0G5L7 A0A1A9W5B1 B5DJ49 A0A224XHI7 A0A182R1M1 A0A0A1WIN6 J3JU08 A0A069DQU1 D1FPK3 A0A3B0JX44 A0A2R7VRE1 J9K4B4 A0A182NNN9 W8CCQ1 A0A0C9QFR4 A0A034VW47 A0A026WX28 A0A1B6L2U0 A0A0K8TWG5 A0A0L7QNE7 A0A195BY33 B4NYP1
A0A2H1W6W5 A0A194PV20 A0A0N1IDE6 A0A212FP47 I4DLD0 A0A067QJN4 A0A1Q3F4E6 A0A1S4F8J4 Q17BZ3 B0X2M5 A0A1B6EQM4 A0A182M893 A0A023EME0 A0A182WXJ1 A0A182GKN4 A0A182Q126 A0A182HQX3 Q5TNW4 A0A182NNP0 A0A182S7A2 A0A182W8E3 A0A182YQV3 A0A1W4WA15 B3N8K0 A0A182PTL4 A0A2M4AER7 B4NYP0 B4MZ14 A0A2M3Z879 A0A1L8EI30 A0A2M4BW00 T1E8C9 A0A336MF19 A0A084VWW1 A0A182FN10 A0A1L8EIB6 B4JE40 B4G7V5 A0A2M3ZI06 B3MPA8 A0A1Y1K631 A0A3B0JU57 Q9VL68 A0A1A9ZVU0 A0A0M3QT57 Q29NE3 A0A0L0CHR4 B4HWC5 A0A1A9V2B2 W5JM77 B4Q896 A0A1A9XJD9 A0A1I8P1X3 A0A1B0BLS1 V5GVQ4 A0A1I8N871 A0A1I8N874 A0A1B6CS95 B4LRD3 U5ESR8 D6WHD7 A0A1W4WXT2 D3TPU1 B4KJE7 A0A0A9WCF2 A0A1B0DDP7 A0A0N7Z960 R4G430 A0A023F9D3 A0A232FMN9 A0A1L8DZ86 A0A0V0G5L7 A0A1A9W5B1 B5DJ49 A0A224XHI7 A0A182R1M1 A0A0A1WIN6 J3JU08 A0A069DQU1 D1FPK3 A0A3B0JX44 A0A2R7VRE1 J9K4B4 A0A182NNN9 W8CCQ1 A0A0C9QFR4 A0A034VW47 A0A026WX28 A0A1B6L2U0 A0A0K8TWG5 A0A0L7QNE7 A0A195BY33 B4NYP1
PDB
1XX4
E-value=2.72606e-51,
Score=509
Ontologies
GO
Topology
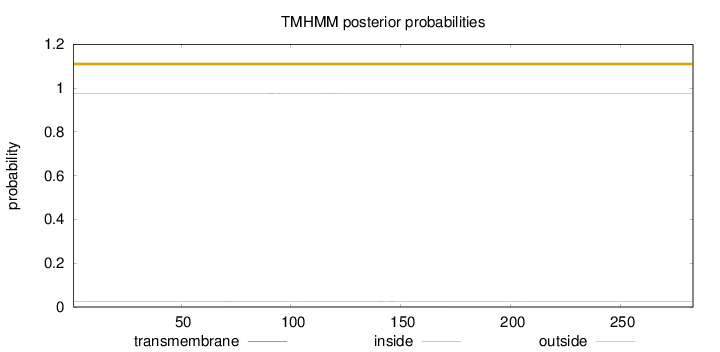
Length:
283
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.04704
Exp number, first 60 AAs:
0.00229
Total prob of N-in:
0.02592
outside
1 - 283
Population Genetic Test Statistics
Pi
17.856736
Theta
157.603315
Tajima's D
0.576217
CLR
0.438676
CSRT
0.532373381330933
Interpretation
Uncertain
