Pre Gene Modal
BGIBMGA011846
Annotation
PREDICTED:_nuclear_envelope_phosphatase-regulatory_subunit_1_homolog_isoform_X2_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 4.049
Sequence
CDS
ATGTCGCTCGAACAAACTGCTTGTGATGATTTGAAAGCTTTTGAGCGTCGTTTAACTGAAGTGATAGCATGTCTCCAACCAGCTACAATGAGATGGAGAATTCTACTGACAATTGTGTCAGTTTGTACAGCAATAGCAGCATACCATTGGTTGATGGATCCTCTGACACCTGTAGTCTCGCTCACACAGTCTTTGTGGAATCATCCCTTTTTTGCTGTGACCTCTACATTATTGGTTTTGCTATTCATGATTGGAGTACATAGAAAGGTAGTGGCTCCTAGCATCATCACAGCGCGCACTCGCTCCATACTTAATGACTTCAACATGTCTTGTGACGATACTGGCAAACTTATTCTGAAGCCGAGACCAGCAAATAGCTCATTATTTTAA
Protein
MSLEQTACDDLKAFERRLTEVIACLQPATMRWRILLTIVSVCTAIAAYHWLMDPLTPVVSLTQSLWNHPFFAVTSTLLVLLFMIGVHRKVVAPSIITARTRSILNDFNMSCDDTGKLILKPRPANSSLF
Summary
Uniprot
H9JQP0
A0A2W1BSW7
A0A2A4J893
A0A2H1W758
S4PGX4
A0A0N1IF38
+ More
A0A3S2M879 A0A212EUL2 A0A194Q1G9 A0A067RF25 A0A1B6KGL0 A0A1B6GZN5 A0A1B6JUI5 A0A0A9X313 A0A087ZTV5 A0A0L7QR06 A0A1B6D043 A0A154PLY2 A0A151IPL5 F4WA11 A0A1W4XWD9 A0A195DL78 A0A158P1C5 A0A0N7Z925 E2C0Q1 A0A026WZH1 A0A2P8Z963 A0A0J7L9M3 E2ATX0 A0A151K0I2 A0A0A9Y6X5 A0A151I2N0 A0A1W4XVH7 A0A158P1C4 N6TDF9 A0A151XH29 A0A0P4WHS1 E9HPY5 A0A0P5FKR7 A0A1B6C3Z3 D6WGC4 A0A146L1Q9 J3JYQ7 A0A0T6AZY7 A0A131X974 A0A023GDT6 G3MN05 A0A3L8DTW0 A0A224YWR2 A0A131Z182 L7M696 A0A1E1WX25 A0A0M8ZXQ6 B7P2W4 V5HDZ1 A0A2R5LF16 Q175U5 A0A023EDS2 B0WT63 A0A087T1K7 A0A224XX37 A0A0V0G8W5 A0A161MLF7 A0A3S3NY71 A0A1Q3F4N3 A0A1L8DKE0 A0A1Q3G1S7 L7LTX7 A0A1B0FC39 A0A1A9Y9J0 A0A1A9VYG7 A0A1B0B9D6 A0A1S3D511 A0A0K8TND7 B4N6Q2 G1N0H6 K7FW56 A0A1D5PB80 H3ATG7 A0A3Q3JXP3 A0A1A9Z3I0 B5X8R3 A0A3B3X6K8 A0A0S7LXP9 M4APW3 A0A087YAV5 A0A3B3U920 A0A3P9NME3 A0A2Y9LMH4 A0A0L0BT94 A0A1D2MKT6 G1P823 A0A1W4UI11 Q6XHR6 I3J401 A0A3Q2DF85 A0A2I4DA22 A0A3Q2ZT27 A0A1A7Z5G9 G3PWW0
A0A3S2M879 A0A212EUL2 A0A194Q1G9 A0A067RF25 A0A1B6KGL0 A0A1B6GZN5 A0A1B6JUI5 A0A0A9X313 A0A087ZTV5 A0A0L7QR06 A0A1B6D043 A0A154PLY2 A0A151IPL5 F4WA11 A0A1W4XWD9 A0A195DL78 A0A158P1C5 A0A0N7Z925 E2C0Q1 A0A026WZH1 A0A2P8Z963 A0A0J7L9M3 E2ATX0 A0A151K0I2 A0A0A9Y6X5 A0A151I2N0 A0A1W4XVH7 A0A158P1C4 N6TDF9 A0A151XH29 A0A0P4WHS1 E9HPY5 A0A0P5FKR7 A0A1B6C3Z3 D6WGC4 A0A146L1Q9 J3JYQ7 A0A0T6AZY7 A0A131X974 A0A023GDT6 G3MN05 A0A3L8DTW0 A0A224YWR2 A0A131Z182 L7M696 A0A1E1WX25 A0A0M8ZXQ6 B7P2W4 V5HDZ1 A0A2R5LF16 Q175U5 A0A023EDS2 B0WT63 A0A087T1K7 A0A224XX37 A0A0V0G8W5 A0A161MLF7 A0A3S3NY71 A0A1Q3F4N3 A0A1L8DKE0 A0A1Q3G1S7 L7LTX7 A0A1B0FC39 A0A1A9Y9J0 A0A1A9VYG7 A0A1B0B9D6 A0A1S3D511 A0A0K8TND7 B4N6Q2 G1N0H6 K7FW56 A0A1D5PB80 H3ATG7 A0A3Q3JXP3 A0A1A9Z3I0 B5X8R3 A0A3B3X6K8 A0A0S7LXP9 M4APW3 A0A087YAV5 A0A3B3U920 A0A3P9NME3 A0A2Y9LMH4 A0A0L0BT94 A0A1D2MKT6 G1P823 A0A1W4UI11 Q6XHR6 I3J401 A0A3Q2DF85 A0A2I4DA22 A0A3Q2ZT27 A0A1A7Z5G9 G3PWW0
Pubmed
19121390
28756777
23622113
26354079
22118469
24845553
+ More
25401762 26823975 21719571 21347285 27129103 20798317 24508170 29403074 23537049 21292972 18362917 19820115 22516182 28049606 22216098 30249741 28797301 26830274 25576852 28503490 25765539 17510324 24945155 26483478 26369729 17994087 20838655 17381049 15592404 9215903 20433749 23542700 26108605 27289101 21993624 14525923 17550304 25186727
25401762 26823975 21719571 21347285 27129103 20798317 24508170 29403074 23537049 21292972 18362917 19820115 22516182 28049606 22216098 30249741 28797301 26830274 25576852 28503490 25765539 17510324 24945155 26483478 26369729 17994087 20838655 17381049 15592404 9215903 20433749 23542700 26108605 27289101 21993624 14525923 17550304 25186727
EMBL
BABH01012183
KZ149984
PZC75736.1
NWSH01002655
PCG67744.1
ODYU01006705
+ More
SOQ48786.1 GAIX01006020 JAA86540.1 KQ461161 KPJ08043.1 RSAL01000010 RVE53692.1 AGBW02012350 OWR45182.1 KQ459590 KPI97245.1 KK852729 KDR17573.1 GEBQ01029379 JAT10598.1 GECZ01001889 JAS67880.1 GECU01004845 JAT02862.1 GBHO01030456 GBHO01025333 GBHO01025331 GBHO01025165 GBRD01005987 GDHC01003772 JAG13148.1 JAG18271.1 JAG18273.1 JAG18439.1 JAG59834.1 JAQ14857.1 KQ414784 KOC61062.1 GEDC01018345 JAS18953.1 KQ434973 KZC12767.1 KQ976836 KYN07971.1 GL888037 EGI68941.1 KQ980762 KYN13592.1 ADTU01006224 GDKW01001731 JAI54864.1 GL451846 EFN78445.1 KK107054 EZA61470.1 PYGN01000141 PSN53037.1 LBMM01000186 KMR04553.1 GL442747 EFN63101.1 KQ981250 KYN44209.1 GBHO01025329 GBHO01025166 GBHO01025164 GBHO01014782 GDHC01020586 JAG18275.1 JAG18438.1 JAG18440.1 JAG28822.1 JAP98042.1 KQ976535 KYM81452.1 APGK01034685 APGK01034686 KB740914 KB632352 ENN78404.1 ERL93284.1 KQ982138 KYQ59667.1 GDRN01049513 JAI66548.1 GL732712 EFX66204.1 GDIQ01256810 GDIQ01254017 LRGB01002190 JAJ97707.1 KZS08737.1 GEDC01029076 GEDC01017934 JAS08222.1 JAS19364.1 KQ971329 EFA01129.2 GDHC01016145 JAQ02484.1 BT128387 AEE63345.1 LJIG01016425 KRT80641.1 GEFH01004497 JAP64084.1 GBBM01003419 JAC31999.1 JO843256 AEO34873.1 QOIP01000004 RLU23603.1 GFPF01010219 MAA21365.1 GEDV01004341 JAP84216.1 GACK01006420 JAA58614.1 GFAC01007599 JAT91589.1 KQ435831 KOX71709.1 ABJB010695952 DS624854 EEC00936.1 GADI01003008 GANP01002643 GEFM01006104 JAA70800.1 JAB81825.1 JAP69692.1 GGLE01003943 MBY08069.1 CH477395 EAT41860.1 JXUM01029023 JXUM01029024 JXUM01098606 JXUM01098607 JXUM01098608 GAPW01005871 GAPW01005870 KQ564434 KQ560839 JAC07728.1 KXJ72286.1 KXJ80666.1 DS232080 EDS34206.1 KK112982 KFM58996.1 GFTR01001958 JAW14468.1 GECL01001580 JAP04544.1 GEMB01002365 JAS00818.1 NCKU01005766 RWS04185.1 GFDL01012520 JAV22525.1 GFDF01007239 JAV06845.1 GFDL01001293 JAV33752.1 GACK01010686 JAA54348.1 CCAG010016063 JXJN01010368 GDAI01001696 JAI15907.1 CH964161 EDW80041.1 AGCU01072717 AGCU01072718 AADN05000558 AFYH01073018 BT047432 ACI67233.1 GBYX01139950 JAO94238.1 AYCK01004723 JRES01001499 KNC22419.1 LJIJ01000964 ODM93521.1 AAPE02057183 AY232117 CM000159 AAR10140.1 EDW94025.1 AERX01000137 HADW01014257 HADX01015364 SBP37596.1
SOQ48786.1 GAIX01006020 JAA86540.1 KQ461161 KPJ08043.1 RSAL01000010 RVE53692.1 AGBW02012350 OWR45182.1 KQ459590 KPI97245.1 KK852729 KDR17573.1 GEBQ01029379 JAT10598.1 GECZ01001889 JAS67880.1 GECU01004845 JAT02862.1 GBHO01030456 GBHO01025333 GBHO01025331 GBHO01025165 GBRD01005987 GDHC01003772 JAG13148.1 JAG18271.1 JAG18273.1 JAG18439.1 JAG59834.1 JAQ14857.1 KQ414784 KOC61062.1 GEDC01018345 JAS18953.1 KQ434973 KZC12767.1 KQ976836 KYN07971.1 GL888037 EGI68941.1 KQ980762 KYN13592.1 ADTU01006224 GDKW01001731 JAI54864.1 GL451846 EFN78445.1 KK107054 EZA61470.1 PYGN01000141 PSN53037.1 LBMM01000186 KMR04553.1 GL442747 EFN63101.1 KQ981250 KYN44209.1 GBHO01025329 GBHO01025166 GBHO01025164 GBHO01014782 GDHC01020586 JAG18275.1 JAG18438.1 JAG18440.1 JAG28822.1 JAP98042.1 KQ976535 KYM81452.1 APGK01034685 APGK01034686 KB740914 KB632352 ENN78404.1 ERL93284.1 KQ982138 KYQ59667.1 GDRN01049513 JAI66548.1 GL732712 EFX66204.1 GDIQ01256810 GDIQ01254017 LRGB01002190 JAJ97707.1 KZS08737.1 GEDC01029076 GEDC01017934 JAS08222.1 JAS19364.1 KQ971329 EFA01129.2 GDHC01016145 JAQ02484.1 BT128387 AEE63345.1 LJIG01016425 KRT80641.1 GEFH01004497 JAP64084.1 GBBM01003419 JAC31999.1 JO843256 AEO34873.1 QOIP01000004 RLU23603.1 GFPF01010219 MAA21365.1 GEDV01004341 JAP84216.1 GACK01006420 JAA58614.1 GFAC01007599 JAT91589.1 KQ435831 KOX71709.1 ABJB010695952 DS624854 EEC00936.1 GADI01003008 GANP01002643 GEFM01006104 JAA70800.1 JAB81825.1 JAP69692.1 GGLE01003943 MBY08069.1 CH477395 EAT41860.1 JXUM01029023 JXUM01029024 JXUM01098606 JXUM01098607 JXUM01098608 GAPW01005871 GAPW01005870 KQ564434 KQ560839 JAC07728.1 KXJ72286.1 KXJ80666.1 DS232080 EDS34206.1 KK112982 KFM58996.1 GFTR01001958 JAW14468.1 GECL01001580 JAP04544.1 GEMB01002365 JAS00818.1 NCKU01005766 RWS04185.1 GFDL01012520 JAV22525.1 GFDF01007239 JAV06845.1 GFDL01001293 JAV33752.1 GACK01010686 JAA54348.1 CCAG010016063 JXJN01010368 GDAI01001696 JAI15907.1 CH964161 EDW80041.1 AGCU01072717 AGCU01072718 AADN05000558 AFYH01073018 BT047432 ACI67233.1 GBYX01139950 JAO94238.1 AYCK01004723 JRES01001499 KNC22419.1 LJIJ01000964 ODM93521.1 AAPE02057183 AY232117 CM000159 AAR10140.1 EDW94025.1 AERX01000137 HADW01014257 HADX01015364 SBP37596.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000283053
UP000007151
UP000053268
+ More
UP000027135 UP000005203 UP000053825 UP000076502 UP000078542 UP000007755 UP000192223 UP000078492 UP000005205 UP000008237 UP000053097 UP000245037 UP000036403 UP000000311 UP000078541 UP000078540 UP000019118 UP000030742 UP000075809 UP000000305 UP000076858 UP000007266 UP000279307 UP000053105 UP000001555 UP000008820 UP000069940 UP000249989 UP000002320 UP000054359 UP000285301 UP000092444 UP000092443 UP000078200 UP000092460 UP000079169 UP000007798 UP000001645 UP000007267 UP000000539 UP000008672 UP000261600 UP000092445 UP000261480 UP000002852 UP000028760 UP000261500 UP000242638 UP000248483 UP000037069 UP000094527 UP000001074 UP000192221 UP000002282 UP000005207 UP000265020 UP000192220 UP000264800 UP000007635
UP000027135 UP000005203 UP000053825 UP000076502 UP000078542 UP000007755 UP000192223 UP000078492 UP000005205 UP000008237 UP000053097 UP000245037 UP000036403 UP000000311 UP000078541 UP000078540 UP000019118 UP000030742 UP000075809 UP000000305 UP000076858 UP000007266 UP000279307 UP000053105 UP000001555 UP000008820 UP000069940 UP000249989 UP000002320 UP000054359 UP000285301 UP000092444 UP000092443 UP000078200 UP000092460 UP000079169 UP000007798 UP000001645 UP000007267 UP000000539 UP000008672 UP000261600 UP000092445 UP000261480 UP000002852 UP000028760 UP000261500 UP000242638 UP000248483 UP000037069 UP000094527 UP000001074 UP000192221 UP000002282 UP000005207 UP000265020 UP000192220 UP000264800 UP000007635
PRIDE
Pfam
PF09771 Tmemb_18A
Interpro
IPR019168
NEP1-R1
ProteinModelPortal
H9JQP0
A0A2W1BSW7
A0A2A4J893
A0A2H1W758
S4PGX4
A0A0N1IF38
+ More
A0A3S2M879 A0A212EUL2 A0A194Q1G9 A0A067RF25 A0A1B6KGL0 A0A1B6GZN5 A0A1B6JUI5 A0A0A9X313 A0A087ZTV5 A0A0L7QR06 A0A1B6D043 A0A154PLY2 A0A151IPL5 F4WA11 A0A1W4XWD9 A0A195DL78 A0A158P1C5 A0A0N7Z925 E2C0Q1 A0A026WZH1 A0A2P8Z963 A0A0J7L9M3 E2ATX0 A0A151K0I2 A0A0A9Y6X5 A0A151I2N0 A0A1W4XVH7 A0A158P1C4 N6TDF9 A0A151XH29 A0A0P4WHS1 E9HPY5 A0A0P5FKR7 A0A1B6C3Z3 D6WGC4 A0A146L1Q9 J3JYQ7 A0A0T6AZY7 A0A131X974 A0A023GDT6 G3MN05 A0A3L8DTW0 A0A224YWR2 A0A131Z182 L7M696 A0A1E1WX25 A0A0M8ZXQ6 B7P2W4 V5HDZ1 A0A2R5LF16 Q175U5 A0A023EDS2 B0WT63 A0A087T1K7 A0A224XX37 A0A0V0G8W5 A0A161MLF7 A0A3S3NY71 A0A1Q3F4N3 A0A1L8DKE0 A0A1Q3G1S7 L7LTX7 A0A1B0FC39 A0A1A9Y9J0 A0A1A9VYG7 A0A1B0B9D6 A0A1S3D511 A0A0K8TND7 B4N6Q2 G1N0H6 K7FW56 A0A1D5PB80 H3ATG7 A0A3Q3JXP3 A0A1A9Z3I0 B5X8R3 A0A3B3X6K8 A0A0S7LXP9 M4APW3 A0A087YAV5 A0A3B3U920 A0A3P9NME3 A0A2Y9LMH4 A0A0L0BT94 A0A1D2MKT6 G1P823 A0A1W4UI11 Q6XHR6 I3J401 A0A3Q2DF85 A0A2I4DA22 A0A3Q2ZT27 A0A1A7Z5G9 G3PWW0
A0A3S2M879 A0A212EUL2 A0A194Q1G9 A0A067RF25 A0A1B6KGL0 A0A1B6GZN5 A0A1B6JUI5 A0A0A9X313 A0A087ZTV5 A0A0L7QR06 A0A1B6D043 A0A154PLY2 A0A151IPL5 F4WA11 A0A1W4XWD9 A0A195DL78 A0A158P1C5 A0A0N7Z925 E2C0Q1 A0A026WZH1 A0A2P8Z963 A0A0J7L9M3 E2ATX0 A0A151K0I2 A0A0A9Y6X5 A0A151I2N0 A0A1W4XVH7 A0A158P1C4 N6TDF9 A0A151XH29 A0A0P4WHS1 E9HPY5 A0A0P5FKR7 A0A1B6C3Z3 D6WGC4 A0A146L1Q9 J3JYQ7 A0A0T6AZY7 A0A131X974 A0A023GDT6 G3MN05 A0A3L8DTW0 A0A224YWR2 A0A131Z182 L7M696 A0A1E1WX25 A0A0M8ZXQ6 B7P2W4 V5HDZ1 A0A2R5LF16 Q175U5 A0A023EDS2 B0WT63 A0A087T1K7 A0A224XX37 A0A0V0G8W5 A0A161MLF7 A0A3S3NY71 A0A1Q3F4N3 A0A1L8DKE0 A0A1Q3G1S7 L7LTX7 A0A1B0FC39 A0A1A9Y9J0 A0A1A9VYG7 A0A1B0B9D6 A0A1S3D511 A0A0K8TND7 B4N6Q2 G1N0H6 K7FW56 A0A1D5PB80 H3ATG7 A0A3Q3JXP3 A0A1A9Z3I0 B5X8R3 A0A3B3X6K8 A0A0S7LXP9 M4APW3 A0A087YAV5 A0A3B3U920 A0A3P9NME3 A0A2Y9LMH4 A0A0L0BT94 A0A1D2MKT6 G1P823 A0A1W4UI11 Q6XHR6 I3J401 A0A3Q2DF85 A0A2I4DA22 A0A3Q2ZT27 A0A1A7Z5G9 G3PWW0
Ontologies
GO
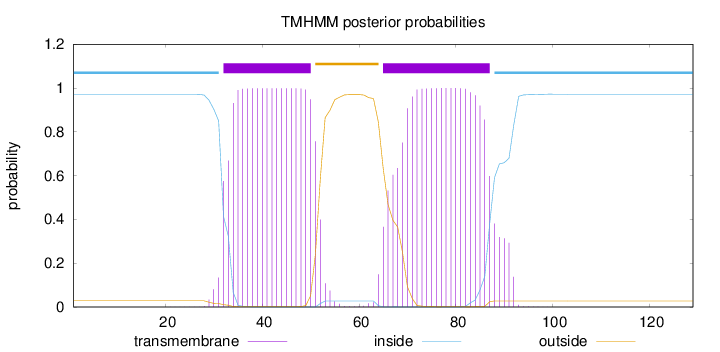
Topology
Length:
129
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
41.44779
Exp number, first 60 AAs:
19.73781
Total prob of N-in:
0.97195
POSSIBLE N-term signal
sequence
inside
1 - 31
TMhelix
32 - 50
outside
51 - 64
TMhelix
65 - 87
inside
88 - 129
Population Genetic Test Statistics
Pi
265.774065
Theta
172.654102
Tajima's D
1.455539
CLR
0.307493
CSRT
0.776161191940403
Interpretation
Uncertain
