Gene
KWMTBOMO06514
Pre Gene Modal
BGIBMGA011634
Annotation
PREDICTED:_C-type_lectin_domain_family_4_member_F_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.282
Sequence
CDS
ATGGCAGAAATAGGCGGGGCTAATTGGTTCAAAGCCCAGCAATACTGCAGGTTCCATGGGATGCACCTCGCTTCCATATCATCGCAACAGGAAAACGATAAACTGGAGCAATACGTTAAGGACTCAGGTTACGGTCGCGAACATTTCTGGACGTCGGGCACTGATTTAGCTGAAGAAGGAAATTTCTTTTGGATGGCGAACGGCAGGCCGCTTACCTTTGTCAATTGGAACGCCGGAGAGCCGAACAACTTCAGATACGAAAATGGCGAAGAAGAGAACTGCTTGGAACTATGGAACAGAGACGATAAAGGCCTCAAATGGAACGATTCCCCTTGTTCGTTCGAAACATACTTCATTTGCGAAGTCCGATCAGATTGA
Protein
MAEIGGANWFKAQQYCRFHGMHLASISSQQENDKLEQYVKDSGYGREHFWTSGTDLAEEGNFFWMANGRPLTFVNWNAGEPNNFRYENGEEENCLELWNRDDKGLKWNDSPCSFETYFICEVRSD
Summary
Uniprot
H9JQ28
A0A3S2NQL0
A0A2W1BQ51
A0A3G1HH47
A0A2A4J7D7
A0A2H1W6S3
+ More
I4DJ91 A0A0N1I522 A0A1E1WHX5 A0A212EUK9 A0A219YXJ8 A0A1B0CV96 A0A1J1I5P3 A0A1A9W4K6 A0A1W4VVQ2 A0A0R3P5W5 B4MXZ3 A0A0K8UG01 A0A034WLM5 W8AQZ3 A0A0J9UBV7 Q9W0I2 A0A1W4W6R7 A0A0R1DZU7 A0A0Q5U1Q2 A0A0Q9XBR9 N6TSW0 A0A034WI61 A0A3B0K9G3 A0A0P9AJR5 Q29EV3 A0A0Q9WU37 A0A0R3P5U4 A0A139W9K8 A0A1W4W855 B4QLZ0 B4HVQ7 B4PD61 A0A0K8V5W1 A0A0A1WZC5 B3NB82 Q8IRH8 B4LG81 A0A1Y1KXD4 D7EJZ5 B3M533 B4J2F0 B4L0N0 A0A139W9J1 A0A3F2YQ66 A0A0K8UZQ3 W5JJD6 A0A182JFN1 A0A1I8P2S6 A0A084VAT5 Q7Q7F9 A0A182N7X9 A0A2C9GWR3 A0A182KEH5 A0A0T6B0G3 A0A023EJ40 A0A182R0N0 A0A0M4EAG2 A0A3F2YQ72 J9HTH4 A0A2C9GWR2 A0A0C9PPN4 A0A0C9QHZ7 A0A182KW05 A0A3F2YSK5 A0A182UY70 A7UTM5 A0A182YPZ5 J9HJ94 A0A1I8N606 A0A2C9GWR9 E2BQT0 F4WBH8 A0A3F2YSK9 A7UTL4 A0A0L7R9D0 A0A195DKI0 A0A154PHZ1 E2AP28 A0A2S2PUU6 A0A2A3E3G9 A0A026WEP8 A0A151WX17 A0A088A5P4 A0A1B0FB62 J9K4D3 A0A232ENA7 A0A195D7A0 A0A195FU96 K7J6I4 A0A023F4S1 A0A2J7R9U3 A0A1B6EW47 A0A182XD36 A0A1B6H0B5
I4DJ91 A0A0N1I522 A0A1E1WHX5 A0A212EUK9 A0A219YXJ8 A0A1B0CV96 A0A1J1I5P3 A0A1A9W4K6 A0A1W4VVQ2 A0A0R3P5W5 B4MXZ3 A0A0K8UG01 A0A034WLM5 W8AQZ3 A0A0J9UBV7 Q9W0I2 A0A1W4W6R7 A0A0R1DZU7 A0A0Q5U1Q2 A0A0Q9XBR9 N6TSW0 A0A034WI61 A0A3B0K9G3 A0A0P9AJR5 Q29EV3 A0A0Q9WU37 A0A0R3P5U4 A0A139W9K8 A0A1W4W855 B4QLZ0 B4HVQ7 B4PD61 A0A0K8V5W1 A0A0A1WZC5 B3NB82 Q8IRH8 B4LG81 A0A1Y1KXD4 D7EJZ5 B3M533 B4J2F0 B4L0N0 A0A139W9J1 A0A3F2YQ66 A0A0K8UZQ3 W5JJD6 A0A182JFN1 A0A1I8P2S6 A0A084VAT5 Q7Q7F9 A0A182N7X9 A0A2C9GWR3 A0A182KEH5 A0A0T6B0G3 A0A023EJ40 A0A182R0N0 A0A0M4EAG2 A0A3F2YQ72 J9HTH4 A0A2C9GWR2 A0A0C9PPN4 A0A0C9QHZ7 A0A182KW05 A0A3F2YSK5 A0A182UY70 A7UTM5 A0A182YPZ5 J9HJ94 A0A1I8N606 A0A2C9GWR9 E2BQT0 F4WBH8 A0A3F2YSK9 A7UTL4 A0A0L7R9D0 A0A195DKI0 A0A154PHZ1 E2AP28 A0A2S2PUU6 A0A2A3E3G9 A0A026WEP8 A0A151WX17 A0A088A5P4 A0A1B0FB62 J9K4D3 A0A232ENA7 A0A195D7A0 A0A195FU96 K7J6I4 A0A023F4S1 A0A2J7R9U3 A0A1B6EW47 A0A182XD36 A0A1B6H0B5
Pubmed
19121390
28756777
22651552
26354079
22118469
15632085
+ More
17994087 25348373 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 23537049 18362917 19820115 25830018 28004739 20920257 23761445 24438588 12364791 24945155 17510324 20966253 14747013 17210077 25244985 25315136 20798317 21719571 24508170 30249741 28648823 20075255 25474469
17994087 25348373 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 23537049 18362917 19820115 25830018 28004739 20920257 23761445 24438588 12364791 24945155 17510324 20966253 14747013 17210077 25244985 25315136 20798317 21719571 24508170 30249741 28648823 20075255 25474469
EMBL
BABH01012190
RSAL01000010
RVE53696.1
KZ149984
PZC75744.1
KY111305
+ More
AQX37241.1 NWSH01002906 PCG67323.1 ODYU01006705 SOQ48781.1 AK401359 KQ459590 BAM17981.1 KPI97242.1 KQ461161 KPJ08038.1 GDQN01004465 JAT86589.1 AGBW02012350 OWR45178.1 KX550119 AQY54436.1 AJWK01030316 AJWK01030317 CVRI01000042 CRK95647.1 CH379070 KRT08514.1 CH963876 EDW76982.1 GDHF01026783 JAI25531.1 GAKP01003720 JAC55232.1 GAMC01015301 GAMC01015300 GAMC01015299 GAMC01015298 JAB91255.1 CM002912 KMY96780.1 AE014296 BT012477 AAF47464.1 AAN11467.1 AAS93748.1 CM000159 KRK00724.1 CH954178 KQS43055.1 CH933809 KRG05984.1 APGK01055668 APGK01055669 KB741269 KB632330 ENN71431.1 ERL92597.1 GAKP01003721 JAC55231.1 OUUW01000009 SPP84760.1 CH902618 KPU78073.1 EAL29957.3 CH940647 KRF84350.1 KRT08513.1 KQ972213 KYB24589.1 CM000363 EDX08776.1 KMY96779.1 CH480817 EDW50022.1 EDW92809.1 GDHF01024635 GDHF01018073 JAI27679.1 JAI34241.1 GBXI01016144 GBXI01010090 GBXI01000584 JAC98147.1 JAD04202.1 JAD13708.1 EDV50135.1 BT004478 AAN11466.1 AAO42642.1 EDW69389.2 GEZM01075601 JAV64046.1 EFA12925.2 EDV39512.2 CH916366 EDV96009.1 EDW18107.2 KYB24590.1 GDHF01020175 JAI32139.1 ADMH02001057 ETN64256.1 ATLV01004459 KE524214 KFB35079.1 AAAB01008960 EAA11819.4 LJIG01016355 KRT80849.1 GAPW01004759 JAC08839.1 AXCN02000907 CP012525 ALC44241.1 CH477779 EJY57928.1 GBYB01003163 JAG72930.1 GBYB01003164 JAG72931.1 APCN01004435 EDO63645.1 EJY57929.1 GL449799 EFN81955.1 GL888063 EGI68457.1 EDO63644.1 KQ414627 KOC67485.1 KQ980765 KYN13356.1 KQ434914 KZC11432.1 GL441439 EFN64860.1 GGMR01020536 MBY33155.1 KZ288400 PBC26255.1 KK107242 QOIP01000001 EZA54542.1 RLU26875.1 KQ982691 KYQ52205.1 CCAG010023727 CCAG010023728 CCAG010023729 ABLF02032234 NNAY01003188 OXU19844.1 KQ976749 KYN08765.1 KQ981272 KYN43867.1 GBBI01002256 JAC16456.1 NEVH01006580 PNF37596.1 GECZ01027641 JAS42128.1 GECZ01004545 GECZ01001653 JAS65224.1 JAS68116.1
AQX37241.1 NWSH01002906 PCG67323.1 ODYU01006705 SOQ48781.1 AK401359 KQ459590 BAM17981.1 KPI97242.1 KQ461161 KPJ08038.1 GDQN01004465 JAT86589.1 AGBW02012350 OWR45178.1 KX550119 AQY54436.1 AJWK01030316 AJWK01030317 CVRI01000042 CRK95647.1 CH379070 KRT08514.1 CH963876 EDW76982.1 GDHF01026783 JAI25531.1 GAKP01003720 JAC55232.1 GAMC01015301 GAMC01015300 GAMC01015299 GAMC01015298 JAB91255.1 CM002912 KMY96780.1 AE014296 BT012477 AAF47464.1 AAN11467.1 AAS93748.1 CM000159 KRK00724.1 CH954178 KQS43055.1 CH933809 KRG05984.1 APGK01055668 APGK01055669 KB741269 KB632330 ENN71431.1 ERL92597.1 GAKP01003721 JAC55231.1 OUUW01000009 SPP84760.1 CH902618 KPU78073.1 EAL29957.3 CH940647 KRF84350.1 KRT08513.1 KQ972213 KYB24589.1 CM000363 EDX08776.1 KMY96779.1 CH480817 EDW50022.1 EDW92809.1 GDHF01024635 GDHF01018073 JAI27679.1 JAI34241.1 GBXI01016144 GBXI01010090 GBXI01000584 JAC98147.1 JAD04202.1 JAD13708.1 EDV50135.1 BT004478 AAN11466.1 AAO42642.1 EDW69389.2 GEZM01075601 JAV64046.1 EFA12925.2 EDV39512.2 CH916366 EDV96009.1 EDW18107.2 KYB24590.1 GDHF01020175 JAI32139.1 ADMH02001057 ETN64256.1 ATLV01004459 KE524214 KFB35079.1 AAAB01008960 EAA11819.4 LJIG01016355 KRT80849.1 GAPW01004759 JAC08839.1 AXCN02000907 CP012525 ALC44241.1 CH477779 EJY57928.1 GBYB01003163 JAG72930.1 GBYB01003164 JAG72931.1 APCN01004435 EDO63645.1 EJY57929.1 GL449799 EFN81955.1 GL888063 EGI68457.1 EDO63644.1 KQ414627 KOC67485.1 KQ980765 KYN13356.1 KQ434914 KZC11432.1 GL441439 EFN64860.1 GGMR01020536 MBY33155.1 KZ288400 PBC26255.1 KK107242 QOIP01000001 EZA54542.1 RLU26875.1 KQ982691 KYQ52205.1 CCAG010023727 CCAG010023728 CCAG010023729 ABLF02032234 NNAY01003188 OXU19844.1 KQ976749 KYN08765.1 KQ981272 KYN43867.1 GBBI01002256 JAC16456.1 NEVH01006580 PNF37596.1 GECZ01027641 JAS42128.1 GECZ01004545 GECZ01001653 JAS65224.1 JAS68116.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000092461 UP000183832 UP000091820 UP000192221 UP000001819 UP000007798 UP000000803 UP000002282 UP000008711 UP000009192 UP000019118 UP000030742 UP000268350 UP000007801 UP000008792 UP000007266 UP000000304 UP000001292 UP000001070 UP000069272 UP000000673 UP000075880 UP000095300 UP000030765 UP000007062 UP000075884 UP000075900 UP000075881 UP000075886 UP000092553 UP000008820 UP000075882 UP000075840 UP000075903 UP000076408 UP000095301 UP000008237 UP000007755 UP000053825 UP000078492 UP000076502 UP000000311 UP000242457 UP000053097 UP000279307 UP000075809 UP000005203 UP000092444 UP000007819 UP000215335 UP000078542 UP000078541 UP000002358 UP000235965
UP000092461 UP000183832 UP000091820 UP000192221 UP000001819 UP000007798 UP000000803 UP000002282 UP000008711 UP000009192 UP000019118 UP000030742 UP000268350 UP000007801 UP000008792 UP000007266 UP000000304 UP000001292 UP000001070 UP000069272 UP000000673 UP000075880 UP000095300 UP000030765 UP000007062 UP000075884 UP000075900 UP000075881 UP000075886 UP000092553 UP000008820 UP000075882 UP000075840 UP000075903 UP000076408 UP000095301 UP000008237 UP000007755 UP000053825 UP000078492 UP000076502 UP000000311 UP000242457 UP000053097 UP000279307 UP000075809 UP000005203 UP000092444 UP000007819 UP000215335 UP000078542 UP000078541 UP000002358 UP000235965
Pfam
PF00059 Lectin_C
Interpro
SUPFAM
SSF56436
SSF56436
Gene 3D
ProteinModelPortal
H9JQ28
A0A3S2NQL0
A0A2W1BQ51
A0A3G1HH47
A0A2A4J7D7
A0A2H1W6S3
+ More
I4DJ91 A0A0N1I522 A0A1E1WHX5 A0A212EUK9 A0A219YXJ8 A0A1B0CV96 A0A1J1I5P3 A0A1A9W4K6 A0A1W4VVQ2 A0A0R3P5W5 B4MXZ3 A0A0K8UG01 A0A034WLM5 W8AQZ3 A0A0J9UBV7 Q9W0I2 A0A1W4W6R7 A0A0R1DZU7 A0A0Q5U1Q2 A0A0Q9XBR9 N6TSW0 A0A034WI61 A0A3B0K9G3 A0A0P9AJR5 Q29EV3 A0A0Q9WU37 A0A0R3P5U4 A0A139W9K8 A0A1W4W855 B4QLZ0 B4HVQ7 B4PD61 A0A0K8V5W1 A0A0A1WZC5 B3NB82 Q8IRH8 B4LG81 A0A1Y1KXD4 D7EJZ5 B3M533 B4J2F0 B4L0N0 A0A139W9J1 A0A3F2YQ66 A0A0K8UZQ3 W5JJD6 A0A182JFN1 A0A1I8P2S6 A0A084VAT5 Q7Q7F9 A0A182N7X9 A0A2C9GWR3 A0A182KEH5 A0A0T6B0G3 A0A023EJ40 A0A182R0N0 A0A0M4EAG2 A0A3F2YQ72 J9HTH4 A0A2C9GWR2 A0A0C9PPN4 A0A0C9QHZ7 A0A182KW05 A0A3F2YSK5 A0A182UY70 A7UTM5 A0A182YPZ5 J9HJ94 A0A1I8N606 A0A2C9GWR9 E2BQT0 F4WBH8 A0A3F2YSK9 A7UTL4 A0A0L7R9D0 A0A195DKI0 A0A154PHZ1 E2AP28 A0A2S2PUU6 A0A2A3E3G9 A0A026WEP8 A0A151WX17 A0A088A5P4 A0A1B0FB62 J9K4D3 A0A232ENA7 A0A195D7A0 A0A195FU96 K7J6I4 A0A023F4S1 A0A2J7R9U3 A0A1B6EW47 A0A182XD36 A0A1B6H0B5
I4DJ91 A0A0N1I522 A0A1E1WHX5 A0A212EUK9 A0A219YXJ8 A0A1B0CV96 A0A1J1I5P3 A0A1A9W4K6 A0A1W4VVQ2 A0A0R3P5W5 B4MXZ3 A0A0K8UG01 A0A034WLM5 W8AQZ3 A0A0J9UBV7 Q9W0I2 A0A1W4W6R7 A0A0R1DZU7 A0A0Q5U1Q2 A0A0Q9XBR9 N6TSW0 A0A034WI61 A0A3B0K9G3 A0A0P9AJR5 Q29EV3 A0A0Q9WU37 A0A0R3P5U4 A0A139W9K8 A0A1W4W855 B4QLZ0 B4HVQ7 B4PD61 A0A0K8V5W1 A0A0A1WZC5 B3NB82 Q8IRH8 B4LG81 A0A1Y1KXD4 D7EJZ5 B3M533 B4J2F0 B4L0N0 A0A139W9J1 A0A3F2YQ66 A0A0K8UZQ3 W5JJD6 A0A182JFN1 A0A1I8P2S6 A0A084VAT5 Q7Q7F9 A0A182N7X9 A0A2C9GWR3 A0A182KEH5 A0A0T6B0G3 A0A023EJ40 A0A182R0N0 A0A0M4EAG2 A0A3F2YQ72 J9HTH4 A0A2C9GWR2 A0A0C9PPN4 A0A0C9QHZ7 A0A182KW05 A0A3F2YSK5 A0A182UY70 A7UTM5 A0A182YPZ5 J9HJ94 A0A1I8N606 A0A2C9GWR9 E2BQT0 F4WBH8 A0A3F2YSK9 A7UTL4 A0A0L7R9D0 A0A195DKI0 A0A154PHZ1 E2AP28 A0A2S2PUU6 A0A2A3E3G9 A0A026WEP8 A0A151WX17 A0A088A5P4 A0A1B0FB62 J9K4D3 A0A232ENA7 A0A195D7A0 A0A195FU96 K7J6I4 A0A023F4S1 A0A2J7R9U3 A0A1B6EW47 A0A182XD36 A0A1B6H0B5
PDB
6BBE
E-value=1.42521e-10,
Score=152
Ontologies
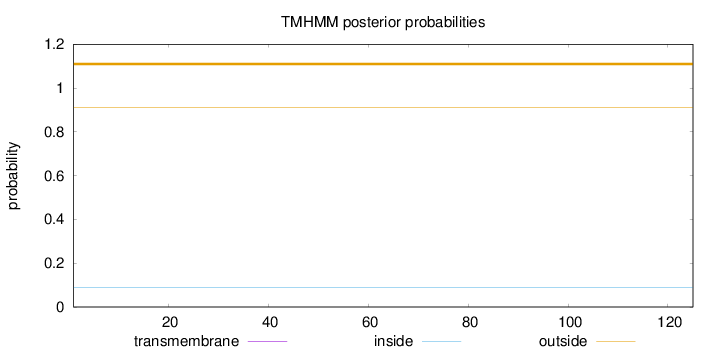
Topology
Length:
125
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00105
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.08857
outside
1 - 125
Population Genetic Test Statistics
Pi
29.56921
Theta
26.288102
Tajima's D
0.230327
CLR
0.93288
CSRT
0.432828358582071
Interpretation
Uncertain
