Gene
KWMTBOMO06502 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011851
Annotation
peritrophin_type-A_domain_protein_4_[Mamestra_configurata]
Location in the cell
Nuclear Reliability : 2.778
Sequence
CDS
ATGAACAAAGCCTTGTGGCTCCTGTGTTGCGCGGCGGCGCTGGTCGGGGCCTCCGCATTCTCAATAAACTTGAGACAGAACCCGGAGACGCTGTGCATTGGCCAGGAGAACTTCCTGAGGATAGCCAGCATTGAAGATTGTTTCCACTACTTCCAGTGCTATGCTGGCCGCTCATATCTTATGCGATGTCCAGAAGAGACATGGTTTAACGAGGCAGAACAGGTTTGCGACTGGTCGGACCTACCAGAGCGCTGTGCAAAAGTTGAACCAGAACCAGAACCAGAGCCGGAACCTGAACCTGAACCCGAACCAGAACCCGAGCCTGAACCTGAGCCGGAACCAGAACCGGAGATAGAACCAGAACAAGAGCCTGAGCCAGAATCTGAACCCGAACTCGAACCCATTGATTCTGAGGAGAACCAAGTAGAAGAACCAGCGCAGCCTAGTGATGAAGTTGCTGAAAACGATGAAGGGTCCGGAGAAGAATGGCAGGAGGATGCCGTTTGGCGTTTTGTTAAATCTTCTCGCGTAAGACAGGAAGAAGTTGAACAAGAAGACGATCAAGACGATGAAGACGAAGAAGGCACTGATACTGTGGAAAATGATAATGATGACAATGAGGAGGATGACGACGAGAATGAGGAAGAGGAAGAAGCTGAAAAACGCTCGAAGAGATCATTGAGACAAGAAGAAACTGAGGGTGATGATGAGAATGAGGATGATGAGGATGAGGACGAGGATGAGGAAGAAGTTGAAAAGCGCTCTAAGAGATCATTGAAACAAGAAGAAACCGAGGATGATGATGATGATGATGATGAGGAGGAAGATGAGGACGAGGAAGAAGTTGAAAAGCGCTCGAAGAGATCATTGAAACAAGAAGAAACCGAGGATGATGATGATGATGATGATGAGGAGGGAGATGAGGACGAGGAAGAAGTTGAAAAGCGCTCGAAGAGATCATTGAAACAAGAAGAAACTGAGGATGATGATGATGATGATGATGAGGAGGATGAAGACGAGGATGAGGAAGAGGAAGAAGCTGAAAAGCGCTCGAAGAGATCATCGAGACAAGAAGAAACCGAGGGTGATGATGACGATGAGGATGATGAGGACGAGGATGAGGAAGAAGTTGAAAAGCGCTCTAAGAGATCATTGAAACAAGAAGAAACTGAGGTTAATGATGACGACGATTATGATGACGAGGATGGTGTCGAAGACGAGGACGAGGAAGAAGTTGAAAAGCGCTCTAAGAGATCATTGAGGCAAGAAGAAACTGAGGATAATGATGACGATGAGGATGAGGATGATGATGATAATGAGGATGATGTCAATGACGAGGACGAGGAAGAAGTTGAAAAGCGCTCTAAGAGATCATTGAGGCAAGAAGAAACAGAAAACGATGATGATGATCAAGAAGAGATTGAAGACGCAGAGGAAGAACTTGAAGCCGAAGAGGAAGAATTAAAAGAAAAGAGCTTCATTAAGAGGTTTTTTAAATTTTTCGCTTGA
Protein
MNKALWLLCCAAALVGASAFSINLRQNPETLCIGQENFLRIASIEDCFHYFQCYAGRSYLMRCPEETWFNEAEQVCDWSDLPERCAKVEPEPEPEPEPEPEPEPEPEPEPEPEPEPEIEPEQEPEPESEPELEPIDSEENQVEEPAQPSDEVAENDEGSGEEWQEDAVWRFVKSSRVRQEEVEQEDDQDDEDEEGTDTVENDNDDNEEDDDENEEEEEAEKRSKRSLRQEETEGDDENEDDEDEDEDEEEVEKRSKRSLKQEETEDDDDDDDEEEDEDEEEVEKRSKRSLKQEETEDDDDDDDEEGDEDEEEVEKRSKRSLKQEETEDDDDDDDEEDEDEDEEEEEAEKRSKRSSRQEETEGDDDDEDDEDEDEEEVEKRSKRSLKQEETEVNDDDDYDDEDGVEDEDEEEVEKRSKRSLRQEETEDNDDDEDEDDDDNEDDVNDEDEEEVEKRSKRSLRQEETENDDDDQEEIEDAEEELEAEEEELKEKSFIKRFFKFFA
Summary
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
Pfam
PF01607 CBM_14
SUPFAM
SSF57625
SSF57625
ProteinModelPortal
PDB
5WVG
E-value=0.00329724,
Score=97
Ontologies
GO
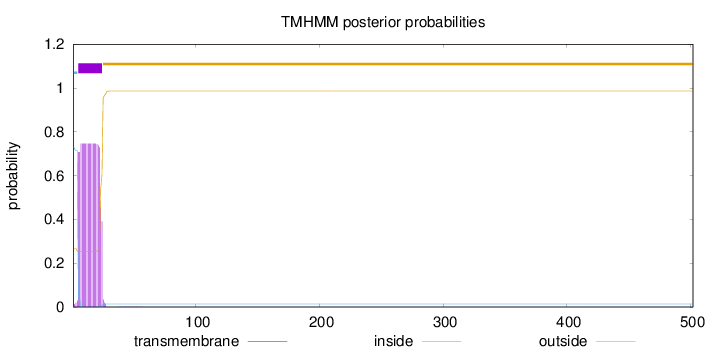
Topology
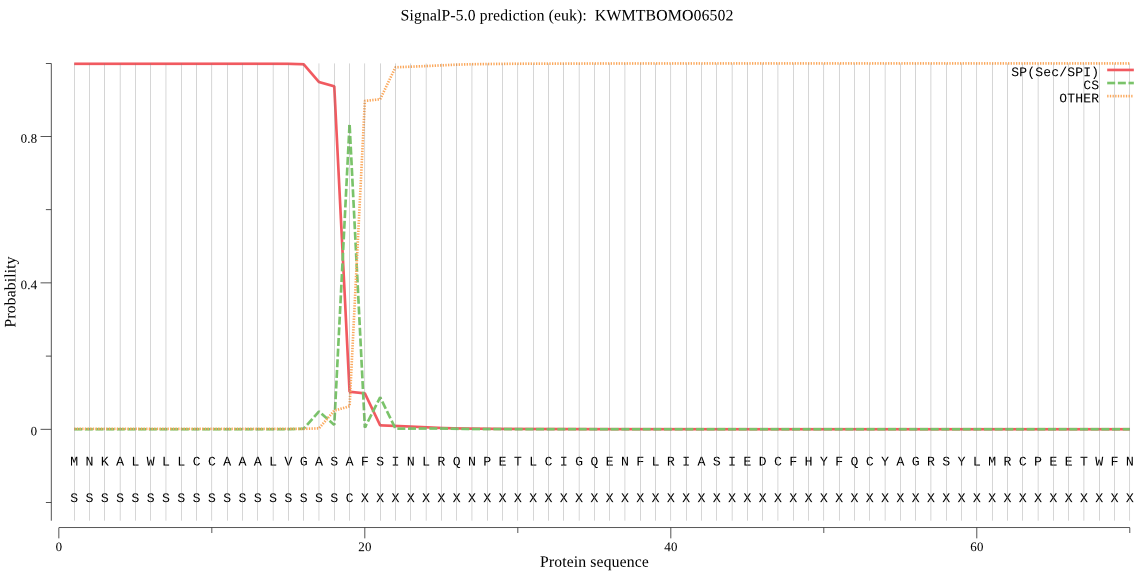
SignalP
Position: 1 - 19,
Likelihood: 0.998802
Length:
502
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
14.30371
Exp number, first 60 AAs:
14.3037
Total prob of N-in:
0.73314
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 24
outside
25 - 502
Population Genetic Test Statistics
Pi
262.930267
Theta
146.469709
Tajima's D
2.258561
CLR
0.016265
CSRT
0.923453827308635
Interpretation
Uncertain
