Gene
KWMTBOMO06494
Pre Gene Modal
BGIBMGA011852
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_protein_cueball_[Bombyx_mori]
Full name
Protein cueball
Location in the cell
Nuclear Reliability : 2.715
Sequence
CDS
ATGGAAAATGAAATTCCCGAAGCGCTCGCCGTGGACAGCTGTAGAGGGTATATTTATTGGACCAATACGAATCTCTCCAGTCCATCGATTGGGAGGGCTAAGTTCGACGGTACAGACAGAAAAACGATCGTTAGCAATATAAGATACACCCCAGTGGCTATAACTTTAGATCAACAGACGAAGAAAATTTACTGGATTAACTCAGAAGGTATCAACTATTCTATTGAAAGCTCAGATTTAGATGGCGCAAACAGGAACATATTACTCACGGCTTCAACGCATCACGACCCTATCGCTATATCGGTGTCCAAGGATTATATTTATTGGATCAGCAAAGCCACTCATTCCATATGGACCCTGCCAAAGAACTCCGATACTTTAACGGATCGCGGTAATGAACTGATCAAATCCAGCAACGAAATAATCGATATATTTGCTCATTACAAAATCGAGGATCAAATCCGTAACATCGAAGAATGTCAACATTTGTCGAATTTGACGGAGAGCAAACGCGTCCAAGAGACGACCACAGCTCTACCGGCTATCGACTACACTGAATTCTGTGAACACGATGCTACAAATGAGAACCGATTCAGTTGCAAATGTCCCGAAGGAATCTATAGGGAGAGATGTGAAGCATTCGTTTGCATGAACTATTGTTTTAATGGAGACTGCACACTCAATAAAGACGGACAACCACAATGCGGATGTAAAGAGGGATACTCCGGTAGAAGATGCGAAATCAATATTTGTCGCGGTTACTGTCTGAACAGCGGTGCCTGTACTGTGGATGATAAACTGAAGCCAATTTGTAAATGTCAGCTAGGACACGAAGGTTCTAGATGTGAAACCGATGTACGGGCCACGACCACAAGCAAAAACATTGAGTCTACTACAAGCACTGTTACTGAAATAGCTAAAACATATAATACTAGTGATTTACCAAGAATCCCGTCCACTTCAAGCGATAATTGCAATTGCACCTCTATCCGGGCGGCCGCTGATGCATTCGAATCGGAAAGGATGGCAAGCGGTGATGGAGAATTCGTAGATTCAGGCGACTCCGGCGTCGATAGAAGCTTAATAGCGTTGGGCGTGATATGCGGGGTGTTGGCCATCACTTGCGTTATGCTCATCACTAAAATCATGCAGCTGACGAAGAGACCAAGAATAAAGAAGCGTTTCATTGTGAACAAGAATGTAACACCAATGACGGCGCGCCCCGACCAGTGCGAGATAACCATCGAGAACTGTTGCAATATGAACATCTGTGAGACGGTCGATGACCCCTGGTCTCCTCGCCAGTAA
Protein
MENEIPEALAVDSCRGYIYWTNTNLSSPSIGRAKFDGTDRKTIVSNIRYTPVAITLDQQTKKIYWINSEGINYSIESSDLDGANRNILLTASTHHDPIAISVSKDYIYWISKATHSIWTLPKNSDTLTDRGNELIKSSNEIIDIFAHYKIEDQIRNIEECQHLSNLTESKRVQETTTALPAIDYTEFCEHDATNENRFSCKCPEGIYRERCEAFVCMNYCFNGDCTLNKDGQPQCGCKEGYSGRRCEINICRGYCLNSGACTVDDKLKPICKCQLGHEGSRCETDVRATTTSKNIESTTSTVTEIAKTYNTSDLPRIPSTSSDNCNCTSIRAAADAFESERMASGDGEFVDSGDSGVDRSLIALGVICGVLAITCVMLITKIMQLTKRPRIKKRFIVNKNVTPMTARPDQCEITIENCCNMNICETVDDPWSPRQ
Summary
Description
Has a role in spermatogenesis and oogenesis.
Similarity
Belongs to the cueball family.
Keywords
Cell membrane
Complete proteome
Differentiation
Disulfide bond
EGF-like domain
Glycoprotein
Membrane
Oogenesis
Reference proteome
Repeat
Signal
Spermatogenesis
Transmembrane
Transmembrane helix
Feature
chain Protein cueball
Uniprot
A0A3S2PK41
A0A1E1WSG6
A0A2W1BE99
A0A0N1PF81
A0A194Q1F5
A0A1E1W8W3
+ More
A0A212ESN2 A0A2H1WKR7 A0A0M8ZXU8 A0A151JR66 F4WX01 A0A2S2NKW4 A0A195F9I1 A0A151WMZ7 A0A2H8TV61 J9JZ35 A0A1Y1N5K7 A0A151IBZ9 A0A232FEX6 A0A2A3E6Z4 A0A158NCF8 U4UKY5 N6SZG3 A0A2A4IWI0 A0A195BWF6 K7IZS3 E2B682 A0A087ZV58 E0VL66 E9INJ0 A0A067QZJ0 A0A1B6HI02 A0A026X3S8 A0A3L8DHB9 E1ZWB3 W8ADQ4 A0A154PNZ9 T1PF89 A0A0A1X763 A0A1L8DJ11 A0A1B6CTF3 D6WHF9 A0A1L8DJ21 A0A0C9PIA9 A0A1L8DJ33 Q29FE9 B4H1F5 A0A1B6E185 B4LCX4 B4IXJ2 A0A0L7R1C4 A0A1B0D1J6 T1GGQ1 A0A1W4X1L9 A0A1I8Q5M8 A0A1B0GJN9 B4L8V5 A0A1B0AVM9 A0A1A9YEB7 A0A1A9V407 A0A1A9ZU32 A0A3B0KEX5 A0A1A9WBR9 A0A0T6BBT3 A0A1B0FMF8 A0A0K8VWQ8 A0A0K8U449 A0A224XMT7 A0A034V882 A0A0K8V1G1 A0A2J7QLX4 A0A023EYV1 A0A336MC24 U5EZR2 A0A1J1IS85 U5EXZ2 A0A0M4EXT2 B4MLE8 Q16YE7 A0A182H3P0 A0A0K8S506 A0A182I6Y5 A0A0A9XS19 A0A146LKL3 A0A0K8S4N0 A0A182WWP9 B0WH58 A0A182Q847 A0A1Q3F7Z9 A0A1Q3F7U3
A0A212ESN2 A0A2H1WKR7 A0A0M8ZXU8 A0A151JR66 F4WX01 A0A2S2NKW4 A0A195F9I1 A0A151WMZ7 A0A2H8TV61 J9JZ35 A0A1Y1N5K7 A0A151IBZ9 A0A232FEX6 A0A2A3E6Z4 A0A158NCF8 U4UKY5 N6SZG3 A0A2A4IWI0 A0A195BWF6 K7IZS3 E2B682 A0A087ZV58 E0VL66 E9INJ0 A0A067QZJ0 A0A1B6HI02 A0A026X3S8 A0A3L8DHB9 E1ZWB3 W8ADQ4 A0A154PNZ9 T1PF89 A0A0A1X763 A0A1L8DJ11 A0A1B6CTF3 D6WHF9 A0A1L8DJ21 A0A0C9PIA9 A0A1L8DJ33 Q29FE9 B4H1F5 A0A1B6E185 B4LCX4 B4IXJ2 A0A0L7R1C4 A0A1B0D1J6 T1GGQ1 A0A1W4X1L9 A0A1I8Q5M8 A0A1B0GJN9 B4L8V5 A0A1B0AVM9 A0A1A9YEB7 A0A1A9V407 A0A1A9ZU32 A0A3B0KEX5 A0A1A9WBR9 A0A0T6BBT3 A0A1B0FMF8 A0A0K8VWQ8 A0A0K8U449 A0A224XMT7 A0A034V882 A0A0K8V1G1 A0A2J7QLX4 A0A023EYV1 A0A336MC24 U5EZR2 A0A1J1IS85 U5EXZ2 A0A0M4EXT2 B4MLE8 Q16YE7 A0A182H3P0 A0A0K8S506 A0A182I6Y5 A0A0A9XS19 A0A146LKL3 A0A0K8S4N0 A0A182WWP9 B0WH58 A0A182Q847 A0A1Q3F7Z9 A0A1Q3F7U3
Pubmed
EMBL
RSAL01000010
RVE53706.1
GDQN01001114
JAT89940.1
KZ150225
PZC72064.1
+ More
KQ461161 KPJ08025.1 KQ459590 KPI97230.1 GDQN01007659 JAT83395.1 AGBW02012761 OWR44498.1 ODYU01009283 SOQ53597.1 KQ435830 KOX71779.1 KQ978619 KYN29764.1 GL888417 EGI61244.1 GGMR01004787 MBY17406.1 KQ981727 KYN37036.1 KQ982934 KYQ49171.1 GFXV01006350 MBW18155.1 ABLF02039830 GEZM01012093 JAV93212.1 KQ978076 KYM97229.1 NNAY01000319 OXU29252.1 KZ288362 PBC26939.1 ADTU01011870 KB632256 ERL90670.1 APGK01050873 KB741178 ENN73164.1 NWSH01005987 PCG63768.1 KQ976396 KYM92917.1 GL445930 EFN88775.1 DS235270 EEB14122.1 GL764306 EFZ17844.1 KK852855 KDR14930.1 GECU01033383 JAS74323.1 KK107034 EZA62044.1 QOIP01000008 RLU19716.1 GL434791 EFN74517.1 GAMC01020285 JAB86270.1 KQ435007 KZC13615.1 KA646563 KA646564 KA647251 AFP61192.1 GBXI01007330 JAD06962.1 GFDF01007641 JAV06443.1 GEDC01030076 GEDC01020479 GEDC01015343 JAS07222.1 JAS16819.1 JAS21955.1 KQ971321 EFA00096.2 GFDF01007642 JAV06442.1 GBYB01000658 GBYB01004049 JAG70425.1 JAG73816.1 GFDF01007643 JAV06441.1 CH379067 EAL31306.2 CH479202 EDW30132.1 GEDC01005618 GEDC01000281 JAS31680.1 JAS37017.1 CH940647 EDW68835.1 CH916366 EDV96429.1 KQ414668 KOC64648.1 AJVK01022074 CAQQ02142924 CAQQ02142925 CAQQ02142926 AJWK01021992 AJWK01021993 CH933816 EDW17130.1 JXJN01004319 JXJN01004320 OUUW01000009 SPP84859.1 LJIG01002138 KRT84802.1 CCAG010014092 GDHF01009003 JAI43311.1 GDHF01031229 JAI21085.1 GFTR01007117 JAW09309.1 GAKP01020630 JAC38322.1 GDHF01019572 GDHF01001089 JAI32742.1 JAI51225.1 NEVH01013240 PNF29591.1 GBBI01004339 JAC14373.1 UFQT01000633 SSX25937.1 GANO01000314 JAB59557.1 CVRI01000057 CRL01966.1 GANO01000664 JAB59207.1 CP012525 ALC42857.1 CH963847 EDW72804.1 CH477517 EAT39657.1 JXUM01107733 JXUM01107734 KQ565169 KXJ71258.1 GBRD01017583 JAG48244.1 APCN01003711 APCN01003712 GBHO01021158 JAG22446.1 GDHC01010824 GDHC01004890 JAQ07805.1 JAQ13739.1 GBRD01017582 GBRD01017581 JAG48246.1 DS231933 EDS27565.1 AXCN02000440 GFDL01011412 JAV23633.1 GFDL01011419 JAV23626.1
KQ461161 KPJ08025.1 KQ459590 KPI97230.1 GDQN01007659 JAT83395.1 AGBW02012761 OWR44498.1 ODYU01009283 SOQ53597.1 KQ435830 KOX71779.1 KQ978619 KYN29764.1 GL888417 EGI61244.1 GGMR01004787 MBY17406.1 KQ981727 KYN37036.1 KQ982934 KYQ49171.1 GFXV01006350 MBW18155.1 ABLF02039830 GEZM01012093 JAV93212.1 KQ978076 KYM97229.1 NNAY01000319 OXU29252.1 KZ288362 PBC26939.1 ADTU01011870 KB632256 ERL90670.1 APGK01050873 KB741178 ENN73164.1 NWSH01005987 PCG63768.1 KQ976396 KYM92917.1 GL445930 EFN88775.1 DS235270 EEB14122.1 GL764306 EFZ17844.1 KK852855 KDR14930.1 GECU01033383 JAS74323.1 KK107034 EZA62044.1 QOIP01000008 RLU19716.1 GL434791 EFN74517.1 GAMC01020285 JAB86270.1 KQ435007 KZC13615.1 KA646563 KA646564 KA647251 AFP61192.1 GBXI01007330 JAD06962.1 GFDF01007641 JAV06443.1 GEDC01030076 GEDC01020479 GEDC01015343 JAS07222.1 JAS16819.1 JAS21955.1 KQ971321 EFA00096.2 GFDF01007642 JAV06442.1 GBYB01000658 GBYB01004049 JAG70425.1 JAG73816.1 GFDF01007643 JAV06441.1 CH379067 EAL31306.2 CH479202 EDW30132.1 GEDC01005618 GEDC01000281 JAS31680.1 JAS37017.1 CH940647 EDW68835.1 CH916366 EDV96429.1 KQ414668 KOC64648.1 AJVK01022074 CAQQ02142924 CAQQ02142925 CAQQ02142926 AJWK01021992 AJWK01021993 CH933816 EDW17130.1 JXJN01004319 JXJN01004320 OUUW01000009 SPP84859.1 LJIG01002138 KRT84802.1 CCAG010014092 GDHF01009003 JAI43311.1 GDHF01031229 JAI21085.1 GFTR01007117 JAW09309.1 GAKP01020630 JAC38322.1 GDHF01019572 GDHF01001089 JAI32742.1 JAI51225.1 NEVH01013240 PNF29591.1 GBBI01004339 JAC14373.1 UFQT01000633 SSX25937.1 GANO01000314 JAB59557.1 CVRI01000057 CRL01966.1 GANO01000664 JAB59207.1 CP012525 ALC42857.1 CH963847 EDW72804.1 CH477517 EAT39657.1 JXUM01107733 JXUM01107734 KQ565169 KXJ71258.1 GBRD01017583 JAG48244.1 APCN01003711 APCN01003712 GBHO01021158 JAG22446.1 GDHC01010824 GDHC01004890 JAQ07805.1 JAQ13739.1 GBRD01017582 GBRD01017581 JAG48246.1 DS231933 EDS27565.1 AXCN02000440 GFDL01011412 JAV23633.1 GFDL01011419 JAV23626.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000007151
UP000053105
UP000078492
+ More
UP000007755 UP000078541 UP000075809 UP000007819 UP000078542 UP000215335 UP000242457 UP000005205 UP000030742 UP000019118 UP000218220 UP000078540 UP000002358 UP000008237 UP000005203 UP000009046 UP000027135 UP000053097 UP000279307 UP000000311 UP000076502 UP000095301 UP000007266 UP000001819 UP000008744 UP000008792 UP000001070 UP000053825 UP000092462 UP000015102 UP000192223 UP000095300 UP000092461 UP000009192 UP000092460 UP000092443 UP000078200 UP000092445 UP000268350 UP000091820 UP000092444 UP000235965 UP000183832 UP000092553 UP000007798 UP000008820 UP000069940 UP000249989 UP000075840 UP000076407 UP000002320 UP000075886
UP000007755 UP000078541 UP000075809 UP000007819 UP000078542 UP000215335 UP000242457 UP000005205 UP000030742 UP000019118 UP000218220 UP000078540 UP000002358 UP000008237 UP000005203 UP000009046 UP000027135 UP000053097 UP000279307 UP000000311 UP000076502 UP000095301 UP000007266 UP000001819 UP000008744 UP000008792 UP000001070 UP000053825 UP000092462 UP000015102 UP000192223 UP000095300 UP000092461 UP000009192 UP000092460 UP000092443 UP000078200 UP000092445 UP000268350 UP000091820 UP000092444 UP000235965 UP000183832 UP000092553 UP000007798 UP000008820 UP000069940 UP000249989 UP000075840 UP000076407 UP000002320 UP000075886
Interpro
SUPFAM
SSF52096
SSF52096
Gene 3D
ProteinModelPortal
A0A3S2PK41
A0A1E1WSG6
A0A2W1BE99
A0A0N1PF81
A0A194Q1F5
A0A1E1W8W3
+ More
A0A212ESN2 A0A2H1WKR7 A0A0M8ZXU8 A0A151JR66 F4WX01 A0A2S2NKW4 A0A195F9I1 A0A151WMZ7 A0A2H8TV61 J9JZ35 A0A1Y1N5K7 A0A151IBZ9 A0A232FEX6 A0A2A3E6Z4 A0A158NCF8 U4UKY5 N6SZG3 A0A2A4IWI0 A0A195BWF6 K7IZS3 E2B682 A0A087ZV58 E0VL66 E9INJ0 A0A067QZJ0 A0A1B6HI02 A0A026X3S8 A0A3L8DHB9 E1ZWB3 W8ADQ4 A0A154PNZ9 T1PF89 A0A0A1X763 A0A1L8DJ11 A0A1B6CTF3 D6WHF9 A0A1L8DJ21 A0A0C9PIA9 A0A1L8DJ33 Q29FE9 B4H1F5 A0A1B6E185 B4LCX4 B4IXJ2 A0A0L7R1C4 A0A1B0D1J6 T1GGQ1 A0A1W4X1L9 A0A1I8Q5M8 A0A1B0GJN9 B4L8V5 A0A1B0AVM9 A0A1A9YEB7 A0A1A9V407 A0A1A9ZU32 A0A3B0KEX5 A0A1A9WBR9 A0A0T6BBT3 A0A1B0FMF8 A0A0K8VWQ8 A0A0K8U449 A0A224XMT7 A0A034V882 A0A0K8V1G1 A0A2J7QLX4 A0A023EYV1 A0A336MC24 U5EZR2 A0A1J1IS85 U5EXZ2 A0A0M4EXT2 B4MLE8 Q16YE7 A0A182H3P0 A0A0K8S506 A0A182I6Y5 A0A0A9XS19 A0A146LKL3 A0A0K8S4N0 A0A182WWP9 B0WH58 A0A182Q847 A0A1Q3F7Z9 A0A1Q3F7U3
A0A212ESN2 A0A2H1WKR7 A0A0M8ZXU8 A0A151JR66 F4WX01 A0A2S2NKW4 A0A195F9I1 A0A151WMZ7 A0A2H8TV61 J9JZ35 A0A1Y1N5K7 A0A151IBZ9 A0A232FEX6 A0A2A3E6Z4 A0A158NCF8 U4UKY5 N6SZG3 A0A2A4IWI0 A0A195BWF6 K7IZS3 E2B682 A0A087ZV58 E0VL66 E9INJ0 A0A067QZJ0 A0A1B6HI02 A0A026X3S8 A0A3L8DHB9 E1ZWB3 W8ADQ4 A0A154PNZ9 T1PF89 A0A0A1X763 A0A1L8DJ11 A0A1B6CTF3 D6WHF9 A0A1L8DJ21 A0A0C9PIA9 A0A1L8DJ33 Q29FE9 B4H1F5 A0A1B6E185 B4LCX4 B4IXJ2 A0A0L7R1C4 A0A1B0D1J6 T1GGQ1 A0A1W4X1L9 A0A1I8Q5M8 A0A1B0GJN9 B4L8V5 A0A1B0AVM9 A0A1A9YEB7 A0A1A9V407 A0A1A9ZU32 A0A3B0KEX5 A0A1A9WBR9 A0A0T6BBT3 A0A1B0FMF8 A0A0K8VWQ8 A0A0K8U449 A0A224XMT7 A0A034V882 A0A0K8V1G1 A0A2J7QLX4 A0A023EYV1 A0A336MC24 U5EZR2 A0A1J1IS85 U5EXZ2 A0A0M4EXT2 B4MLE8 Q16YE7 A0A182H3P0 A0A0K8S506 A0A182I6Y5 A0A0A9XS19 A0A146LKL3 A0A0K8S4N0 A0A182WWP9 B0WH58 A0A182Q847 A0A1Q3F7Z9 A0A1Q3F7U3
PDB
3S8Z
E-value=2.54971e-16,
Score=209
Ontologies
GO
Topology
Subcellular location
Cell membrane
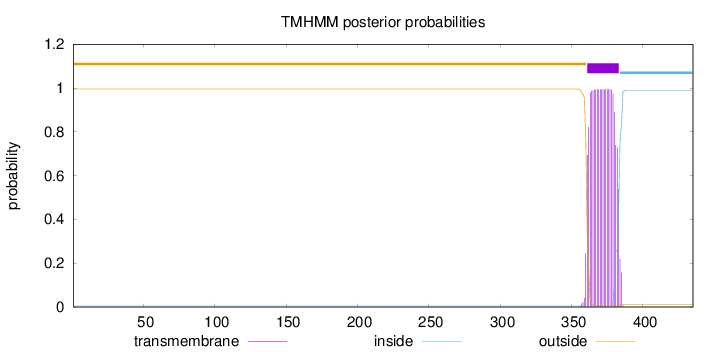
Length:
435
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.97404
Exp number, first 60 AAs:
0.00034
Total prob of N-in:
0.00454
outside
1 - 360
TMhelix
361 - 383
inside
384 - 435
Population Genetic Test Statistics
Pi
159.523982
Theta
142.718938
Tajima's D
0.151261
CLR
0.1456
CSRT
0.415579221038948
Interpretation
Uncertain
