Gene
KWMTBOMO06493 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011625
Annotation
PREDICTED:_85/88_kDa_calcium-independent_phospholipase_A2_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.047
Sequence
CDS
ATGTATTTTGGTCCGGCGGCCGGCGGCGGCGACGAGTCGGCGGCCCCCCCGCCCCCGCCCCCCCACCAGCAGCTCGTGTGGCAGGTGGCGCGCGCCACCGGGGCCGCGCCCTCCTACTTCCGAGCTTCGGGCAGGTACCTGGACGGTGGGCTCATGGGCAACAACCCCACCCTGGACGCGCTCACGGAGCTGGCGGAGCTGGGCCTGGCGCTGCGCGGCACCGGCCGGGGCGAGCTCGCCGCCGCCTGCGGCCTCAAGATAGTCGTGTCCTGCGGGACAGGACAAATCCCCGTGACGCTCGTGAAGGACTTCGACGTGTTCAAGCCGGAGAGCCTGTGGGACACGGCGCGGCTGGCCTGGGGACTGTCGGCGCTGGGCAGCCTGCTGGTCGACCAGGCGACGCAGGCGGACGGGCGCGTGGTGGAGCGGGCGCGCGCGTGGTGCGGCGCGGCGGGCGTGCCCTACTACCGGTTCTCGCCGCCGATGTCGCGCGACGTCAGCATGGACGAGCGCTCCGACGAGAGGCTGGTCTGCATGCTGTGGGAGGCGCAGGCCTACATGCGCGACCACCGGGACGAGGTCGCCGAGCTGGCCGCCCTCCTCGCCGACGCCGGTGAAAATGACAAGTAA
Protein
MYFGPAAGGGDESAAPPPPPPHQQLVWQVARATGAAPSYFRASGRYLDGGLMGNNPTLDALTELAELGLALRGTGRGELAAACGLKIVVSCGTGQIPVTLVKDFDVFKPESLWDTARLAWGLSALGSLLVDQATQADGRVVERARAWCGAAGVPYYRFSPPMSRDVSMDERSDERLVCMLWEAQAYMRDHRDEVAELAALLADAGENDK
Summary
Uniprot
H9JQ19
A0A0N1INE0
A0A2A4JPU5
A0A088MGW0
A0A2H1WE56
A0A3S2NL63
+ More
A0A2W1BK66 A0A3S2M889 A0A0L0CDY4 A0A1B0DI55 B4HAD6 A0A0R3P4Y6 A0A3B0KU46 A0A1Y1M836 B5DQ54 A0A3B0KLJ5 A0A0P9C2F9 B3MA59 A0A0Q9XR36 B4L922 A0A1I8P3D0 A0A0M5IYR7 A0A0A1X4K5 A0A0Q9WTL8 A0A336LVD1 B4LCC4 A0A1I8MIG2 A0A1I8MIG4 T1PCV3 B4N485 Q8MR13 A0A0Q5UIP3 A0A0R1DX12 B3NGN0 B4PDX4 A0A1W4U9F9 Q9VT60 A0A1W4UKP5 A0A2J7RGS8 Q29R25 Q7KUD4 A0A1B0FGI1 A0A0J9RU07 B4HLQ7 B4QNQ4 A0A1A9YNM1 A0A1B0B941 A0A1A9Z341 A0A034VJU9 A0A1A9VAT2 B4J226 A0A182HB36 A0A336LYC0 A0A0P6IYJ3 A0A2M4BCT5 A0A2M3Z5E7 N6U371 A0A023ERJ7 U4UL57 A0A1A9WG44 W8C5L8 A0A182FDJ2 A0A182LRU3 A0A182WCR4 A0A182NNH0 A0A067R7U3 T1DK33 A0A182RKL6 A0A1Q3FC18 A0A1Q3FCF9 A0A2M4A3T4 A0A2M4A3V4 A0A1L8DJ70 A0A2M4A7H0 A0A182QI25 A0A1B0F046 N6U5V5 A0A182PNX4 A0A0N1IU18 W5JEY2 E9IAV4 A0A154P1G8 A0A182K414 Q7Q2U1 A0A182XCQ4 A0A182TDP1 A0A182VHP0 A0A1W4WFT9 A0A182HXM7 A0A182LI21 U5EWQ4 A0A195F645 A0A195EJ46 A0A158NK26 A0A195BV61 A0A151WTK5 A0A1J1J373 F4X1K3 A0A182SAI4 A0A0J7L0J5
A0A2W1BK66 A0A3S2M889 A0A0L0CDY4 A0A1B0DI55 B4HAD6 A0A0R3P4Y6 A0A3B0KU46 A0A1Y1M836 B5DQ54 A0A3B0KLJ5 A0A0P9C2F9 B3MA59 A0A0Q9XR36 B4L922 A0A1I8P3D0 A0A0M5IYR7 A0A0A1X4K5 A0A0Q9WTL8 A0A336LVD1 B4LCC4 A0A1I8MIG2 A0A1I8MIG4 T1PCV3 B4N485 Q8MR13 A0A0Q5UIP3 A0A0R1DX12 B3NGN0 B4PDX4 A0A1W4U9F9 Q9VT60 A0A1W4UKP5 A0A2J7RGS8 Q29R25 Q7KUD4 A0A1B0FGI1 A0A0J9RU07 B4HLQ7 B4QNQ4 A0A1A9YNM1 A0A1B0B941 A0A1A9Z341 A0A034VJU9 A0A1A9VAT2 B4J226 A0A182HB36 A0A336LYC0 A0A0P6IYJ3 A0A2M4BCT5 A0A2M3Z5E7 N6U371 A0A023ERJ7 U4UL57 A0A1A9WG44 W8C5L8 A0A182FDJ2 A0A182LRU3 A0A182WCR4 A0A182NNH0 A0A067R7U3 T1DK33 A0A182RKL6 A0A1Q3FC18 A0A1Q3FCF9 A0A2M4A3T4 A0A2M4A3V4 A0A1L8DJ70 A0A2M4A7H0 A0A182QI25 A0A1B0F046 N6U5V5 A0A182PNX4 A0A0N1IU18 W5JEY2 E9IAV4 A0A154P1G8 A0A182K414 Q7Q2U1 A0A182XCQ4 A0A182TDP1 A0A182VHP0 A0A1W4WFT9 A0A182HXM7 A0A182LI21 U5EWQ4 A0A195F645 A0A195EJ46 A0A158NK26 A0A195BV61 A0A151WTK5 A0A1J1J373 F4X1K3 A0A182SAI4 A0A0J7L0J5
Pubmed
19121390
26354079
28756777
26108605
17994087
15632085
+ More
28004739 25830018 25315136 17550304 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 26483478 26999592 23537049 24945155 24495485 24845553 20920257 23761445 21282665 12364791 20966253 21347285 21719571
28004739 25830018 25315136 17550304 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 26483478 26999592 23537049 24945155 24495485 24845553 20920257 23761445 21282665 12364791 20966253 21347285 21719571
EMBL
BABH01012256
BABH01012257
BABH01012258
BABH01012259
BABH01012260
BABH01012261
+ More
KQ461161 KPJ08024.1 NWSH01000855 PCG73839.1 KJ995815 AIN39484.1 ODYU01008038 SOQ51307.1 RSAL01000010 RVE53707.1 KZ150225 PZC72063.1 RVE53708.1 JRES01000522 KNC30446.1 AJVK01062013 CH479240 EDW37534.1 CH379069 KRT08174.1 OUUW01000012 SPP87448.1 GEZM01037773 JAV81942.1 EDY73721.1 SPP87449.1 CH902618 KPU77849.1 EDV39073.1 CH933816 KRG07668.1 EDW17197.1 CP012525 ALC44081.1 GBXI01008684 JAD05608.1 CH940647 KRF84011.1 UFQT01000224 SSX22002.1 EDW68769.1 KA645788 AFP60417.1 CH964095 EDW78959.2 AY122192 AAM52704.1 CH954178 KQS43751.1 CM000159 KRK01581.1 EDV51266.1 KQS43752.1 EDW93970.1 AE014296 BT125951 AAF50194.3 ADX35930.1 NEVH01003752 PNF40045.1 BT024215 ABC86277.1 AAN11936.2 AAN11937.2 AAN11938.2 CCAG010013633 CM002912 KMY98784.1 CH480815 EDW40939.1 CM000363 EDX09921.1 KMY98783.1 JXJN01010264 GAKP01016555 JAC42397.1 CH916366 EDV95951.1 JXUM01124117 JXUM01124118 JXUM01124119 JXUM01124120 KQ566793 KXJ69824.1 SSX22001.1 GDUN01000021 JAN95898.1 GGFJ01001714 MBW50855.1 GGFM01002973 MBW23724.1 APGK01041340 KB740993 ENN76020.1 GAPW01002299 JAC11299.1 KB632256 ERL90755.1 GAMC01008984 GAMC01008981 JAB97574.1 AXCM01005571 KK852649 KDR19488.1 GAMD01001151 JAB00440.1 GFDL01009920 JAV25125.1 GFDL01009862 JAV25183.1 GGFK01002153 MBW35474.1 GGFK01002173 MBW35494.1 GFDF01007700 JAV06384.1 GGFK01003413 MBW36734.1 AXCN02001910 AJWK01034104 AJWK01034105 AJWK01034106 ENN76021.1 KQ435714 KOX79194.1 ADMH02001698 ETN61384.1 GL762073 EFZ22306.1 KQ434796 KZC05683.1 AAAB01008968 EAA13225.3 APCN01005322 GANO01000453 JAB59418.1 KQ981805 KYN35539.1 KQ978801 KYN28285.1 ADTU01002051 KQ976403 KYM92145.1 KQ982757 KYQ51121.1 CVRI01000064 CRL05329.1 GL888531 EGI59661.1 LBMM01001525 KMQ96141.1
KQ461161 KPJ08024.1 NWSH01000855 PCG73839.1 KJ995815 AIN39484.1 ODYU01008038 SOQ51307.1 RSAL01000010 RVE53707.1 KZ150225 PZC72063.1 RVE53708.1 JRES01000522 KNC30446.1 AJVK01062013 CH479240 EDW37534.1 CH379069 KRT08174.1 OUUW01000012 SPP87448.1 GEZM01037773 JAV81942.1 EDY73721.1 SPP87449.1 CH902618 KPU77849.1 EDV39073.1 CH933816 KRG07668.1 EDW17197.1 CP012525 ALC44081.1 GBXI01008684 JAD05608.1 CH940647 KRF84011.1 UFQT01000224 SSX22002.1 EDW68769.1 KA645788 AFP60417.1 CH964095 EDW78959.2 AY122192 AAM52704.1 CH954178 KQS43751.1 CM000159 KRK01581.1 EDV51266.1 KQS43752.1 EDW93970.1 AE014296 BT125951 AAF50194.3 ADX35930.1 NEVH01003752 PNF40045.1 BT024215 ABC86277.1 AAN11936.2 AAN11937.2 AAN11938.2 CCAG010013633 CM002912 KMY98784.1 CH480815 EDW40939.1 CM000363 EDX09921.1 KMY98783.1 JXJN01010264 GAKP01016555 JAC42397.1 CH916366 EDV95951.1 JXUM01124117 JXUM01124118 JXUM01124119 JXUM01124120 KQ566793 KXJ69824.1 SSX22001.1 GDUN01000021 JAN95898.1 GGFJ01001714 MBW50855.1 GGFM01002973 MBW23724.1 APGK01041340 KB740993 ENN76020.1 GAPW01002299 JAC11299.1 KB632256 ERL90755.1 GAMC01008984 GAMC01008981 JAB97574.1 AXCM01005571 KK852649 KDR19488.1 GAMD01001151 JAB00440.1 GFDL01009920 JAV25125.1 GFDL01009862 JAV25183.1 GGFK01002153 MBW35474.1 GGFK01002173 MBW35494.1 GFDF01007700 JAV06384.1 GGFK01003413 MBW36734.1 AXCN02001910 AJWK01034104 AJWK01034105 AJWK01034106 ENN76021.1 KQ435714 KOX79194.1 ADMH02001698 ETN61384.1 GL762073 EFZ22306.1 KQ434796 KZC05683.1 AAAB01008968 EAA13225.3 APCN01005322 GANO01000453 JAB59418.1 KQ981805 KYN35539.1 KQ978801 KYN28285.1 ADTU01002051 KQ976403 KYM92145.1 KQ982757 KYQ51121.1 CVRI01000064 CRL05329.1 GL888531 EGI59661.1 LBMM01001525 KMQ96141.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000283053
UP000037069
UP000092462
+ More
UP000008744 UP000001819 UP000268350 UP000007801 UP000009192 UP000095300 UP000092553 UP000008792 UP000095301 UP000007798 UP000008711 UP000002282 UP000192221 UP000000803 UP000235965 UP000092444 UP000001292 UP000000304 UP000092443 UP000092460 UP000092445 UP000078200 UP000001070 UP000069940 UP000249989 UP000019118 UP000030742 UP000091820 UP000069272 UP000075883 UP000075920 UP000075884 UP000027135 UP000075900 UP000075886 UP000092461 UP000075885 UP000053105 UP000000673 UP000076502 UP000075881 UP000007062 UP000076407 UP000075902 UP000075903 UP000192223 UP000075840 UP000075882 UP000078541 UP000078492 UP000005205 UP000078540 UP000075809 UP000183832 UP000007755 UP000075901 UP000036403
UP000008744 UP000001819 UP000268350 UP000007801 UP000009192 UP000095300 UP000092553 UP000008792 UP000095301 UP000007798 UP000008711 UP000002282 UP000192221 UP000000803 UP000235965 UP000092444 UP000001292 UP000000304 UP000092443 UP000092460 UP000092445 UP000078200 UP000001070 UP000069940 UP000249989 UP000019118 UP000030742 UP000091820 UP000069272 UP000075883 UP000075920 UP000075884 UP000027135 UP000075900 UP000075886 UP000092461 UP000075885 UP000053105 UP000000673 UP000076502 UP000075881 UP000007062 UP000076407 UP000075902 UP000075903 UP000192223 UP000075840 UP000075882 UP000078541 UP000078492 UP000005205 UP000078540 UP000075809 UP000183832 UP000007755 UP000075901 UP000036403
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JQ19
A0A0N1INE0
A0A2A4JPU5
A0A088MGW0
A0A2H1WE56
A0A3S2NL63
+ More
A0A2W1BK66 A0A3S2M889 A0A0L0CDY4 A0A1B0DI55 B4HAD6 A0A0R3P4Y6 A0A3B0KU46 A0A1Y1M836 B5DQ54 A0A3B0KLJ5 A0A0P9C2F9 B3MA59 A0A0Q9XR36 B4L922 A0A1I8P3D0 A0A0M5IYR7 A0A0A1X4K5 A0A0Q9WTL8 A0A336LVD1 B4LCC4 A0A1I8MIG2 A0A1I8MIG4 T1PCV3 B4N485 Q8MR13 A0A0Q5UIP3 A0A0R1DX12 B3NGN0 B4PDX4 A0A1W4U9F9 Q9VT60 A0A1W4UKP5 A0A2J7RGS8 Q29R25 Q7KUD4 A0A1B0FGI1 A0A0J9RU07 B4HLQ7 B4QNQ4 A0A1A9YNM1 A0A1B0B941 A0A1A9Z341 A0A034VJU9 A0A1A9VAT2 B4J226 A0A182HB36 A0A336LYC0 A0A0P6IYJ3 A0A2M4BCT5 A0A2M3Z5E7 N6U371 A0A023ERJ7 U4UL57 A0A1A9WG44 W8C5L8 A0A182FDJ2 A0A182LRU3 A0A182WCR4 A0A182NNH0 A0A067R7U3 T1DK33 A0A182RKL6 A0A1Q3FC18 A0A1Q3FCF9 A0A2M4A3T4 A0A2M4A3V4 A0A1L8DJ70 A0A2M4A7H0 A0A182QI25 A0A1B0F046 N6U5V5 A0A182PNX4 A0A0N1IU18 W5JEY2 E9IAV4 A0A154P1G8 A0A182K414 Q7Q2U1 A0A182XCQ4 A0A182TDP1 A0A182VHP0 A0A1W4WFT9 A0A182HXM7 A0A182LI21 U5EWQ4 A0A195F645 A0A195EJ46 A0A158NK26 A0A195BV61 A0A151WTK5 A0A1J1J373 F4X1K3 A0A182SAI4 A0A0J7L0J5
A0A2W1BK66 A0A3S2M889 A0A0L0CDY4 A0A1B0DI55 B4HAD6 A0A0R3P4Y6 A0A3B0KU46 A0A1Y1M836 B5DQ54 A0A3B0KLJ5 A0A0P9C2F9 B3MA59 A0A0Q9XR36 B4L922 A0A1I8P3D0 A0A0M5IYR7 A0A0A1X4K5 A0A0Q9WTL8 A0A336LVD1 B4LCC4 A0A1I8MIG2 A0A1I8MIG4 T1PCV3 B4N485 Q8MR13 A0A0Q5UIP3 A0A0R1DX12 B3NGN0 B4PDX4 A0A1W4U9F9 Q9VT60 A0A1W4UKP5 A0A2J7RGS8 Q29R25 Q7KUD4 A0A1B0FGI1 A0A0J9RU07 B4HLQ7 B4QNQ4 A0A1A9YNM1 A0A1B0B941 A0A1A9Z341 A0A034VJU9 A0A1A9VAT2 B4J226 A0A182HB36 A0A336LYC0 A0A0P6IYJ3 A0A2M4BCT5 A0A2M3Z5E7 N6U371 A0A023ERJ7 U4UL57 A0A1A9WG44 W8C5L8 A0A182FDJ2 A0A182LRU3 A0A182WCR4 A0A182NNH0 A0A067R7U3 T1DK33 A0A182RKL6 A0A1Q3FC18 A0A1Q3FCF9 A0A2M4A3T4 A0A2M4A3V4 A0A1L8DJ70 A0A2M4A7H0 A0A182QI25 A0A1B0F046 N6U5V5 A0A182PNX4 A0A0N1IU18 W5JEY2 E9IAV4 A0A154P1G8 A0A182K414 Q7Q2U1 A0A182XCQ4 A0A182TDP1 A0A182VHP0 A0A1W4WFT9 A0A182HXM7 A0A182LI21 U5EWQ4 A0A195F645 A0A195EJ46 A0A158NK26 A0A195BV61 A0A151WTK5 A0A1J1J373 F4X1K3 A0A182SAI4 A0A0J7L0J5
PDB
6AUN
E-value=7.55396e-34,
Score=356
Ontologies
PATHWAY
00564
Glycerophospholipid metabolism - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00590 Arachidonic acid metabolism - Bombyx mori (domestic silkworm)
00591 Linoleic acid metabolism - Bombyx mori (domestic silkworm)
00592 alpha-Linolenic acid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
00565 Ether lipid metabolism - Bombyx mori (domestic silkworm)
00590 Arachidonic acid metabolism - Bombyx mori (domestic silkworm)
00591 Linoleic acid metabolism - Bombyx mori (domestic silkworm)
00592 alpha-Linolenic acid metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
GO
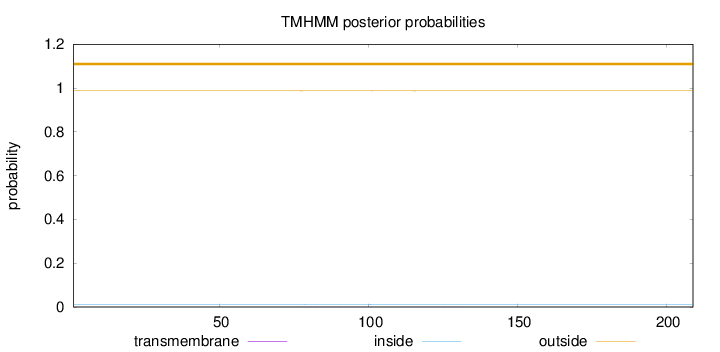
Topology
Length:
209
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02651
Exp number, first 60 AAs:
0.00122
Total prob of N-in:
0.01257
outside
1 - 209
Population Genetic Test Statistics
Pi
47.262801
Theta
144.329991
Tajima's D
-1.152366
CLR
1197.184609
CSRT
0.110644467776611
Interpretation
Uncertain
