Gene
KWMTBOMO06492 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011625
Annotation
PLA2-C_[Spodoptera_exigua]
Location in the cell
Cytoplasmic Reliability : 1.499 Mitochondrial Reliability : 1.682
Sequence
CDS
ATGCTGAATTCATTTTTTCGTGGTCTTCTTCGTACCGAAGAGCTGCCGACCAAAGTCCTCGACGTGCGATCAGACAACTACATCTCACGACCAGTTCACTACAGGGAAGATAGTATGTTATTGTATGGTCCGAAAATACCGAATGACGGTAAAAATCCTAAAGATAAATATTACGAAATCGTTCTATACAAACCGTTTACAGAGAATTTACATCAAATGTACAGTTTACTTCGGTCAGAAACTCTTGAGAGTGCAGAAGAAAAGTTCATAGTGTTCAAAGAAAGAATACCCATTTTTATACAAATTACCAAAGATTGTACCGTTTCAACCCTACAAAAATTATGCGATATTTTGTCGGAGCATCAGTCGTGGACAATAGCGCATTTGATAGCGCACTTTGGCCTGTATGAACACTTGACTCATGCGGCGGTAAAGAAACACATCAATGAAATCGATCCAATCACGGGAGCCACGCCTTTGATGGTGGCCCTGAAGACATCGTGCTTGCGAGTCGTACAATGTTTGGTGTCGCATAATTGCTCTCTGGACACGATTGATGTTGATGGGAACACAGTCTTTCACTACGCGGCAGCTAGCAACAAGGAGATAATTAATATCCTGGCCAGTAAAACGTCTGCGCCTCTGAACGTGTACAACCACCAAGGCTACACGCCGCTGCACATGGCTTGCCTCGCGGACGCGCCTGACTGCGTGCGCGCCCTCATGCTGGCTGGCGCCGATGTTAACCTCTCCGCCGCCAAACGTTCCACAAGCACCTTACCGGGTATCGTGGGCGACCTGCTGCAAGACAATCAGCCCAAGCTGTACCAGCAGGACATGAAGCACGGCGGCACCCCGCTGCACTGGGCCGTCTCCAGGGAGGTCATCGACGCGTTGGTCGACAAGAACTGCGACATCAACGCTTTGAACTTCGACGGCCGCACCGCCCTGCACGTGATGGTGCTGCGCGGCCGGCTCGAGTGTGCGATCGCGCTGCTGTCCAGGGGCGCGGAGATATCTGTGGCCGACAACGAGGGAAACACGCCGCTTCATTCGGCGGTCAAACAATCGTACGTGATCACTCAAGCGCTGGTGGTCTTCGGTGCTGACCTGAACGCGAGGAACAACGCCGGCCACACGCCCAGGCACGCCGTGCTCGCAGACACCTCCAATAACTCTGACAAGATTCTCTATGTTCTGCATTCCGTCGGAGCTAAAAGGTGTCCCCCCGAGCAGGTGGGCTGCGGGCCGGGCTGCGCGGCGCGCGGGGACTACAACGGGGTGCCGCCCTCCACGCTCAGCCCCCCGCCCGCCCGGACGCTCATTAACCACATGCTGTCCGTGGCCGCCATGGAGAAGGCGTCCTGCTTTGATCCCAGCGAAAAACAGGGCCGTTTGCTGTGCCTGGACGGCGGCGGCATCAGGGGCCTGGTGCTCGTCCAACTGCTCCTGAGCCTGGAGGAGGCGGTCGGGAAGCCCATCATACACTGCTTCGACTGGGTGGCCGGGACCAGCACCGGGGGCATCCTGGCGTTGGCGTTGGCCAGCGGGAAAACGTTAAGGGAATGCCAGCAGCTGTACTTCAGACTGAAGGAGCTCGCGTTCGTTGGCATGAGACCGTACCCTTCGGAGGCGCTCGAGAACATCTTGAAGGAGTGCTTAGGTACAGAAACAGTAATGTCGGATATAAAGCATCCGAAGCTGATGATCCTGGCCGTGCTCGCCGACCGGAAGCCGGTGGACCTGCACATCTTCAGGAACTACAAGTCCGCTCATGAAATACTCAGCGAACACAACGGCACGACCGGTTAG
Protein
MLNSFFRGLLRTEELPTKVLDVRSDNYISRPVHYREDSMLLYGPKIPNDGKNPKDKYYEIVLYKPFTENLHQMYSLLRSETLESAEEKFIVFKERIPIFIQITKDCTVSTLQKLCDILSEHQSWTIAHLIAHFGLYEHLTHAAVKKHINEIDPITGATPLMVALKTSCLRVVQCLVSHNCSLDTIDVDGNTVFHYAAASNKEIINILASKTSAPLNVYNHQGYTPLHMACLADAPDCVRALMLAGADVNLSAAKRSTSTLPGIVGDLLQDNQPKLYQQDMKHGGTPLHWAVSREVIDALVDKNCDINALNFDGRTALHVMVLRGRLECAIALLSRGAEISVADNEGNTPLHSAVKQSYVITQALVVFGADLNARNNAGHTPRHAVLADTSNNSDKILYVLHSVGAKRCPPEQVGCGPGCAARGDYNGVPPSTLSPPPARTLINHMLSVAAMEKASCFDPSEKQGRLLCLDGGGIRGLVLVQLLLSLEEAVGKPIIHCFDWVAGTSTGGILALALASGKTLRECQQLYFRLKELAFVGMRPYPSEALENILKECLGTETVMSDIKHPKLMILAVLADRKPVDLHIFRNYKSAHEILSEHNGTTG
Summary
Uniprot
H9JQ19
A0A2A4JPU5
A0A088MGW0
A0A2W1BK66
A0A0N1INE0
A0A194PWU1
+ More
A0A212ESQ1 A0A3S2M889 A0A2D3E1D1 A0A0L7LDY7 A0A0T6AVS0 A0A3S2NL63 A0A1Y1M836 E9IAV4 A0A2J7RGS8 A0A2J7RGT5 A0A2J7RGV2 D6WG42 E2B1P4 F4X1K3 A0A195F645 A0A158NK26 A0A195EJ46 A0A195BV61 E2BS24 A0A232F9X4 A0A1W4WFT9 A0A067R7U3 A0A151I8T5 A0A151WTK5 A0A154P1G8 A0A2A3ENU8 A0A087ZSA8 K7IS76 A0A026VZC4 A0A0N1IU18 N6U371 A0A0C9QTL1 U4UL57 A0A0C9RF78 A0A310SU08 A0A0L7RDB1 A0A023F206 A0A336LZK7 K7IS75 A0A1B6JAL6 A0A1B6DBA0 J9JZ64 A0A0A9YX16 A0A1B6GC83 A0A336LYC0 E0W0Y8 A0A0A9YZF0 A0A224XGX9 A0A0J7L0J5 N6U5V5 A0A2S2P7Q4 A0A1L8DJ70 A0A0N7ZB75 V5G453 T1IEG7 A0A293LVA7 A0A131XTE3 A0A131YWB0 A0A336LVD1 L7M3H1 A0A147BSG3 A0A2R5LAD4 A0A131YU93 A0A0P5UD31 A0A0P5TQE6 A0A0N8E1M2 A0A0P4Y9A6 A0A0P5AWI5 A0A0P4YY77 A0A0N8D644 A0A0P5MG24 A0A0P5MB37 A0A0P5V4W6 A0A0P5UZG8 A0A0P5V5K9 A0A0P5WSD3 A0A0N8D894 A0A0P5W3L7 A0A0P5LTR0 A0A0P6I0Z2 A0A1B0F046 A0A0N8CT95 A0A0P5YE66 A0A0P5C487 A0A0P5RCW8 A0A1A9WG44 A0A1S4FXB5 Q16KY2 A0A0P6IYJ3 A0A0P6BPB3
A0A212ESQ1 A0A3S2M889 A0A2D3E1D1 A0A0L7LDY7 A0A0T6AVS0 A0A3S2NL63 A0A1Y1M836 E9IAV4 A0A2J7RGS8 A0A2J7RGT5 A0A2J7RGV2 D6WG42 E2B1P4 F4X1K3 A0A195F645 A0A158NK26 A0A195EJ46 A0A195BV61 E2BS24 A0A232F9X4 A0A1W4WFT9 A0A067R7U3 A0A151I8T5 A0A151WTK5 A0A154P1G8 A0A2A3ENU8 A0A087ZSA8 K7IS76 A0A026VZC4 A0A0N1IU18 N6U371 A0A0C9QTL1 U4UL57 A0A0C9RF78 A0A310SU08 A0A0L7RDB1 A0A023F206 A0A336LZK7 K7IS75 A0A1B6JAL6 A0A1B6DBA0 J9JZ64 A0A0A9YX16 A0A1B6GC83 A0A336LYC0 E0W0Y8 A0A0A9YZF0 A0A224XGX9 A0A0J7L0J5 N6U5V5 A0A2S2P7Q4 A0A1L8DJ70 A0A0N7ZB75 V5G453 T1IEG7 A0A293LVA7 A0A131XTE3 A0A131YWB0 A0A336LVD1 L7M3H1 A0A147BSG3 A0A2R5LAD4 A0A131YU93 A0A0P5UD31 A0A0P5TQE6 A0A0N8E1M2 A0A0P4Y9A6 A0A0P5AWI5 A0A0P4YY77 A0A0N8D644 A0A0P5MG24 A0A0P5MB37 A0A0P5V4W6 A0A0P5UZG8 A0A0P5V5K9 A0A0P5WSD3 A0A0N8D894 A0A0P5W3L7 A0A0P5LTR0 A0A0P6I0Z2 A0A1B0F046 A0A0N8CT95 A0A0P5YE66 A0A0P5C487 A0A0P5RCW8 A0A1A9WG44 A0A1S4FXB5 Q16KY2 A0A0P6IYJ3 A0A0P6BPB3
Pubmed
EMBL
BABH01012256
BABH01012257
BABH01012258
BABH01012259
BABH01012260
BABH01012261
+ More
NWSH01000855 PCG73839.1 KJ995815 AIN39484.1 KZ150225 PZC72063.1 KQ461161 KPJ08024.1 KQ459590 KPI97229.1 AGBW02012761 OWR44499.1 RSAL01000010 RVE53708.1 MF443094 ATU31391.1 JTDY01001516 KOB73677.1 LJIG01022694 KRT79208.1 RVE53707.1 GEZM01037773 JAV81942.1 GL762073 EFZ22306.1 NEVH01003752 PNF40045.1 PNF40046.1 PNF40047.1 KQ971319 EFA00525.2 GL444953 EFN60400.1 GL888531 EGI59661.1 KQ981805 KYN35539.1 ADTU01002051 KQ978801 KYN28285.1 KQ976403 KYM92145.1 GL450107 EFN81540.1 NNAY01000598 OXU27475.1 KK852649 KDR19488.1 KQ978331 KYM95078.1 KQ982757 KYQ51121.1 KQ434796 KZC05683.1 KZ288204 PBC33184.1 KK107570 QOIP01000005 EZA48831.1 RLU23023.1 KQ435714 KOX79194.1 APGK01041340 KB740993 ENN76020.1 GBYB01007059 JAG76826.1 KB632256 ERL90755.1 GBYB01007060 JAG76827.1 KQ760382 OAD60715.1 KQ414615 KOC68741.1 GBBI01003329 JAC15383.1 UFQT01000224 SSX22003.1 GECU01011551 JAS96155.1 GEDC01014317 GEDC01008401 JAS22981.1 JAS28897.1 ABLF02027959 GBHO01006047 GBRD01007217 GDHC01004653 JAG37557.1 JAG58604.1 JAQ13976.1 GECZ01009734 JAS60035.1 SSX22001.1 DS235863 EEB19293.1 GBHO01006045 GBRD01007216 GDHC01019538 JAG37559.1 JAG58605.1 JAP99090.1 GFTR01008594 JAW07832.1 LBMM01001525 KMQ96141.1 ENN76021.1 GGMR01012864 MBY25483.1 GFDF01007700 JAV06384.1 GDRN01088379 GDRN01088378 GDRN01088377 JAI60864.1 GALX01003651 JAB64815.1 ACPB03014090 ACPB03014091 GFWV01007266 MAA31996.1 GEFM01005879 JAP69917.1 GEDV01005757 JAP82800.1 SSX22002.1 GACK01007351 JAA57683.1 GEGO01002069 JAR93335.1 GGLE01002336 MBY06462.1 GEDV01005758 JAP82799.1 GDIP01114792 JAL88922.1 GDIP01123074 JAL80640.1 GDIQ01070692 JAN24045.1 GDIP01233024 JAI90377.1 GDIP01193624 JAJ29778.1 GDIP01223068 JAJ00334.1 GDIP01065095 JAM38620.1 GDIQ01159959 JAK91766.1 GDIQ01158162 JAK93563.1 GDIP01104040 LRGB01002101 JAL99674.1 KZS09174.1 GDIP01106637 JAL97077.1 GDIP01104041 JAL99673.1 GDIP01082081 JAM21634.1 GDIP01059095 JAM44620.1 GDIP01091580 JAM12135.1 GDIQ01165752 JAK85973.1 GDIQ01011114 JAN83623.1 AJWK01034104 AJWK01034105 AJWK01034106 GDIP01101087 JAM02628.1 GDIP01059096 JAM44619.1 GDIP01179972 JAJ43430.1 GDIQ01102575 JAL49151.1 CH477932 EAT34967.1 GDUN01000021 JAN95898.1 GDIP01012229 JAM91486.1
NWSH01000855 PCG73839.1 KJ995815 AIN39484.1 KZ150225 PZC72063.1 KQ461161 KPJ08024.1 KQ459590 KPI97229.1 AGBW02012761 OWR44499.1 RSAL01000010 RVE53708.1 MF443094 ATU31391.1 JTDY01001516 KOB73677.1 LJIG01022694 KRT79208.1 RVE53707.1 GEZM01037773 JAV81942.1 GL762073 EFZ22306.1 NEVH01003752 PNF40045.1 PNF40046.1 PNF40047.1 KQ971319 EFA00525.2 GL444953 EFN60400.1 GL888531 EGI59661.1 KQ981805 KYN35539.1 ADTU01002051 KQ978801 KYN28285.1 KQ976403 KYM92145.1 GL450107 EFN81540.1 NNAY01000598 OXU27475.1 KK852649 KDR19488.1 KQ978331 KYM95078.1 KQ982757 KYQ51121.1 KQ434796 KZC05683.1 KZ288204 PBC33184.1 KK107570 QOIP01000005 EZA48831.1 RLU23023.1 KQ435714 KOX79194.1 APGK01041340 KB740993 ENN76020.1 GBYB01007059 JAG76826.1 KB632256 ERL90755.1 GBYB01007060 JAG76827.1 KQ760382 OAD60715.1 KQ414615 KOC68741.1 GBBI01003329 JAC15383.1 UFQT01000224 SSX22003.1 GECU01011551 JAS96155.1 GEDC01014317 GEDC01008401 JAS22981.1 JAS28897.1 ABLF02027959 GBHO01006047 GBRD01007217 GDHC01004653 JAG37557.1 JAG58604.1 JAQ13976.1 GECZ01009734 JAS60035.1 SSX22001.1 DS235863 EEB19293.1 GBHO01006045 GBRD01007216 GDHC01019538 JAG37559.1 JAG58605.1 JAP99090.1 GFTR01008594 JAW07832.1 LBMM01001525 KMQ96141.1 ENN76021.1 GGMR01012864 MBY25483.1 GFDF01007700 JAV06384.1 GDRN01088379 GDRN01088378 GDRN01088377 JAI60864.1 GALX01003651 JAB64815.1 ACPB03014090 ACPB03014091 GFWV01007266 MAA31996.1 GEFM01005879 JAP69917.1 GEDV01005757 JAP82800.1 SSX22002.1 GACK01007351 JAA57683.1 GEGO01002069 JAR93335.1 GGLE01002336 MBY06462.1 GEDV01005758 JAP82799.1 GDIP01114792 JAL88922.1 GDIP01123074 JAL80640.1 GDIQ01070692 JAN24045.1 GDIP01233024 JAI90377.1 GDIP01193624 JAJ29778.1 GDIP01223068 JAJ00334.1 GDIP01065095 JAM38620.1 GDIQ01159959 JAK91766.1 GDIQ01158162 JAK93563.1 GDIP01104040 LRGB01002101 JAL99674.1 KZS09174.1 GDIP01106637 JAL97077.1 GDIP01104041 JAL99673.1 GDIP01082081 JAM21634.1 GDIP01059095 JAM44620.1 GDIP01091580 JAM12135.1 GDIQ01165752 JAK85973.1 GDIQ01011114 JAN83623.1 AJWK01034104 AJWK01034105 AJWK01034106 GDIP01101087 JAM02628.1 GDIP01059096 JAM44619.1 GDIP01179972 JAJ43430.1 GDIQ01102575 JAL49151.1 CH477932 EAT34967.1 GDUN01000021 JAN95898.1 GDIP01012229 JAM91486.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000283053
+ More
UP000037510 UP000235965 UP000007266 UP000000311 UP000007755 UP000078541 UP000005205 UP000078492 UP000078540 UP000008237 UP000215335 UP000192223 UP000027135 UP000078542 UP000075809 UP000076502 UP000242457 UP000005203 UP000002358 UP000053097 UP000279307 UP000053105 UP000019118 UP000030742 UP000053825 UP000007819 UP000009046 UP000036403 UP000015103 UP000076858 UP000092461 UP000091820 UP000008820
UP000037510 UP000235965 UP000007266 UP000000311 UP000007755 UP000078541 UP000005205 UP000078492 UP000078540 UP000008237 UP000215335 UP000192223 UP000027135 UP000078542 UP000075809 UP000076502 UP000242457 UP000005203 UP000002358 UP000053097 UP000279307 UP000053105 UP000019118 UP000030742 UP000053825 UP000007819 UP000009046 UP000036403 UP000015103 UP000076858 UP000092461 UP000091820 UP000008820
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JQ19
A0A2A4JPU5
A0A088MGW0
A0A2W1BK66
A0A0N1INE0
A0A194PWU1
+ More
A0A212ESQ1 A0A3S2M889 A0A2D3E1D1 A0A0L7LDY7 A0A0T6AVS0 A0A3S2NL63 A0A1Y1M836 E9IAV4 A0A2J7RGS8 A0A2J7RGT5 A0A2J7RGV2 D6WG42 E2B1P4 F4X1K3 A0A195F645 A0A158NK26 A0A195EJ46 A0A195BV61 E2BS24 A0A232F9X4 A0A1W4WFT9 A0A067R7U3 A0A151I8T5 A0A151WTK5 A0A154P1G8 A0A2A3ENU8 A0A087ZSA8 K7IS76 A0A026VZC4 A0A0N1IU18 N6U371 A0A0C9QTL1 U4UL57 A0A0C9RF78 A0A310SU08 A0A0L7RDB1 A0A023F206 A0A336LZK7 K7IS75 A0A1B6JAL6 A0A1B6DBA0 J9JZ64 A0A0A9YX16 A0A1B6GC83 A0A336LYC0 E0W0Y8 A0A0A9YZF0 A0A224XGX9 A0A0J7L0J5 N6U5V5 A0A2S2P7Q4 A0A1L8DJ70 A0A0N7ZB75 V5G453 T1IEG7 A0A293LVA7 A0A131XTE3 A0A131YWB0 A0A336LVD1 L7M3H1 A0A147BSG3 A0A2R5LAD4 A0A131YU93 A0A0P5UD31 A0A0P5TQE6 A0A0N8E1M2 A0A0P4Y9A6 A0A0P5AWI5 A0A0P4YY77 A0A0N8D644 A0A0P5MG24 A0A0P5MB37 A0A0P5V4W6 A0A0P5UZG8 A0A0P5V5K9 A0A0P5WSD3 A0A0N8D894 A0A0P5W3L7 A0A0P5LTR0 A0A0P6I0Z2 A0A1B0F046 A0A0N8CT95 A0A0P5YE66 A0A0P5C487 A0A0P5RCW8 A0A1A9WG44 A0A1S4FXB5 Q16KY2 A0A0P6IYJ3 A0A0P6BPB3
A0A212ESQ1 A0A3S2M889 A0A2D3E1D1 A0A0L7LDY7 A0A0T6AVS0 A0A3S2NL63 A0A1Y1M836 E9IAV4 A0A2J7RGS8 A0A2J7RGT5 A0A2J7RGV2 D6WG42 E2B1P4 F4X1K3 A0A195F645 A0A158NK26 A0A195EJ46 A0A195BV61 E2BS24 A0A232F9X4 A0A1W4WFT9 A0A067R7U3 A0A151I8T5 A0A151WTK5 A0A154P1G8 A0A2A3ENU8 A0A087ZSA8 K7IS76 A0A026VZC4 A0A0N1IU18 N6U371 A0A0C9QTL1 U4UL57 A0A0C9RF78 A0A310SU08 A0A0L7RDB1 A0A023F206 A0A336LZK7 K7IS75 A0A1B6JAL6 A0A1B6DBA0 J9JZ64 A0A0A9YX16 A0A1B6GC83 A0A336LYC0 E0W0Y8 A0A0A9YZF0 A0A224XGX9 A0A0J7L0J5 N6U5V5 A0A2S2P7Q4 A0A1L8DJ70 A0A0N7ZB75 V5G453 T1IEG7 A0A293LVA7 A0A131XTE3 A0A131YWB0 A0A336LVD1 L7M3H1 A0A147BSG3 A0A2R5LAD4 A0A131YU93 A0A0P5UD31 A0A0P5TQE6 A0A0N8E1M2 A0A0P4Y9A6 A0A0P5AWI5 A0A0P4YY77 A0A0N8D644 A0A0P5MG24 A0A0P5MB37 A0A0P5V4W6 A0A0P5UZG8 A0A0P5V5K9 A0A0P5WSD3 A0A0N8D894 A0A0P5W3L7 A0A0P5LTR0 A0A0P6I0Z2 A0A1B0F046 A0A0N8CT95 A0A0P5YE66 A0A0P5C487 A0A0P5RCW8 A0A1A9WG44 A0A1S4FXB5 Q16KY2 A0A0P6IYJ3 A0A0P6BPB3
PDB
6AUN
E-value=1.91276e-79,
Score=755
Ontologies
GO
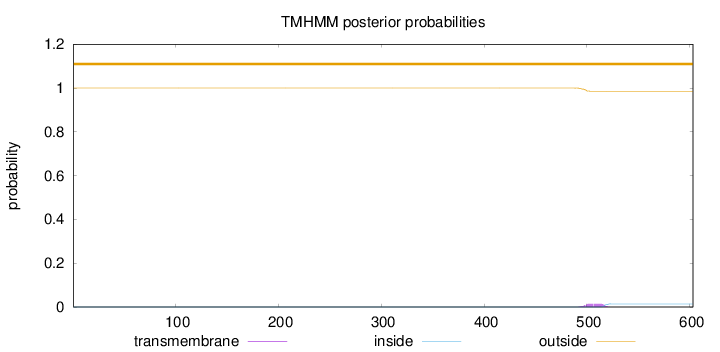
Topology
Length:
603
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29164
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00007
outside
1 - 603
Population Genetic Test Statistics
Pi
245.479482
Theta
193.653509
Tajima's D
1.116024
CLR
0.026393
CSRT
0.690965451727414
Interpretation
Uncertain
