Gene
KWMTBOMO06490
Pre Gene Modal
BGIBMGA011854
Annotation
PREDICTED:_cyclin-dependent_kinase_14_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.15
Sequence
CDS
ATGAGAGAGAAAAGAGGCGGCGCTATGAGCAAAATGCAAAAGCTTCGAAAGCGCCTGTCCTTGAGCTTCGGAAGGTTATCAACGAAAGACGAGTCGGACGGCGACAACGGTGAATGTCGAGGGCGACAGCAGAACGGCGGGGCCCGGGGGAAGCTCTCTTACAATGGATACTCTGAAGAGTGTCTCGATCGACTGGAGCCTAACGGAAATATACCTAATGATAAGGACTCCCATTACGAGTGGAGCGAAGGTGGCAACGGTGAATACCGTGTCCGAAGACAGTTGTCGGTCTCTTCAGATTCGAAGCTCCTGGACGAAGGTGCGAGGGAGGAGGCGCGAGTTGTCATGAGGCCGAAGCGACCTCCCAGACCCAAGAGTGAAGCGTTCCTCGGGCCTCACGAGCAGTCCAGTAGACGCACCAAGCGATTTAGTGCCTTTGGGGGTGACTCTCCGTTCGGTAAATCCGAAGCTTACATTAAACTGGAGCAGCTCGGTGAGGGTTCCTACGCTACAGTATATAAAGGTTATAGCAATTTAACGCAACAAGTGGTAGCACTCAAAGAGATCAGACTGCAGGAGGAGGAAGGCGCGCCCTTCACCGCGATAAGGGAAGCCTCGCTCCTCAAAGAGTTGAAACACGCGAACATAGTCACGTTACACGATATAGTTCACACTAGGGAGACCCTGACGTTCGTCTTCGAGTTTGTGGATACGGATCTTTCACAATACATGGAGAGGCATCCCGGAGGACTGAATCATCACAACGTTAGGCTATTCATGTTCCAACTGCTGAGGGGGCTCTCGTATTGTCATCGGAGAAGAGTATTGCACAGGGACGTGAAGCCGCAGAACCTGTTGATAAGCTCCCAGGGCGAGCTGAAGCTGGCCGACTTCGGTCTGGCGCGGGCCAAGTCGGTCCCGAGCCACACGTACTCGCACGAAGTGGTCACGCTATGGTATCGACCACCAGACGTTTTATTAGGTAGCACGGAGTATTCGACGTCGCTGGACATGTGGGGCGTGGGCTGCATATTCGTGGAGATGATCTGCGGCGTGCCGACGTTTCCCGGCATCAGGGACACTCACGACCAGCTCGATAAAATATTCAAGGTGGTGGGTACGCCCACGGAGGAGAGCTGGTCCGGCGTGTCCCGGCTGCCGGGCGCGCGCGTGCACCTGTCCCGCGGCCCCTCCCCCGCGCGCCCCCTCAGCGCGGCGTTCCCGCTGCTGCGCGAGGCGGGGCGCGACGCCGAGCGGCTGGCGGCGGCGCTGCTGCAGCCCGACCCCGCGCGGCGGCTGCCTGCGCACCGGGCCATACACCACGCCTACTTCGCATCCATACCGCCCAGGATACGGGACCTGCCCGACGAGGTGTCGATCTTCACCGTGGAGGGCGTGTGCCTGCACGCGGAGGCCCGCCACGGCGCCGTCAAGTGA
Protein
MREKRGGAMSKMQKLRKRLSLSFGRLSTKDESDGDNGECRGRQQNGGARGKLSYNGYSEECLDRLEPNGNIPNDKDSHYEWSEGGNGEYRVRRQLSVSSDSKLLDEGAREEARVVMRPKRPPRPKSEAFLGPHEQSSRRTKRFSAFGGDSPFGKSEAYIKLEQLGEGSYATVYKGYSNLTQQVVALKEIRLQEEEGAPFTAIREASLLKELKHANIVTLHDIVHTRETLTFVFEFVDTDLSQYMERHPGGLNHHNVRLFMFQLLRGLSYCHRRRVLHRDVKPQNLLISSQGELKLADFGLARAKSVPSHTYSHEVVTLWYRPPDVLLGSTEYSTSLDMWGVGCIFVEMICGVPTFPGIRDTHDQLDKIFKVVGTPTEESWSGVSRLPGARVHLSRGPSPARPLSAAFPLLREAGRDAERLAAALLQPDPARRLPAHRAIHHAYFASIPPRIRDLPDEVSIFTVEGVCLHAEARHGAVK
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
A0A2W1BCC2
A0A212EIR5
A0A194PW53
H9JQP8
A0A0N1I517
A0A158NA41
+ More
A0A087ZZD4 A0A151WWM6 A0A154PJU9 F4WC24 A0A195DMQ6 E2A5M1 A0A195FR63 A0A310SG71 A0A026VUM0 A0A0P6IXS9 A0A0L7R0Y2 A0A023ESY4 E9IXM1 E2B884 A0A158NA40 A0A336KT93 A0A0N0BIA3 A0A158NA39 A0A195CUX5 A0A0C9R7I1 A0A1Q3FWP3 T1E210 A0A084VPY1 A0A2Y9D0S9 Q7Q6Q3 A0A2M3YZN6 A0A2M4A6R6 A0A2M4CTE2 A0A0C9RAV3 A0A1B6MTZ5 A0A0A9Y8F5 A0A1B6MJS1 A0A0A9W940 D6WH49 A0A139WK61 A0A2A3EIF9 A0A0K8SJ70 A0A146LJT8 A0A1J1ITG9 A0A0K8TR83 U5EUK7 J9JWI5 A0A2S2N6T1 A0A1W4WFH7 A0A2H8TE41 A0A1W4WF71 A0A1Y1L9M3 T1D3Y2 A0A1B6LLF0 A0A146LWS3 A0A1B6LNE6 A0A2M4A6Y6 A0A2S2QD51 A0A1L8DWX7 B0WMQ5 A0A195BKC8 A0A1L8DKM5 V5I9C0 A0A182Q7T8 A0A067R6P1 A0A1I8MY59 A0A1W4W041 A0A1W4VNV0 A0A3B0K3J1 A0A0J9RN84 Q7KM05 Q7KM08 Q94888 A0A3B0KPW9 A0A3B0KL04 A0A3B0JYW5 A0A1I8MY72 A0A1I8MY64 C8VUZ5 B4PH45 A0A023F4Q8 A0A1I8PTX4 A0A0J9RNB4 Q7KM03 A0A1W4W0Y2 B4LDI5 C9QP46 B3NBX7 A0A3B0K1Q0 M9PE01 A0A1W4W0X8 B4QPK1 B4KYC0 A0A3B0KTL8 B4N572 B4HTS8 Q9XTK9 A0A0J9RNS1 Q7KM04
A0A087ZZD4 A0A151WWM6 A0A154PJU9 F4WC24 A0A195DMQ6 E2A5M1 A0A195FR63 A0A310SG71 A0A026VUM0 A0A0P6IXS9 A0A0L7R0Y2 A0A023ESY4 E9IXM1 E2B884 A0A158NA40 A0A336KT93 A0A0N0BIA3 A0A158NA39 A0A195CUX5 A0A0C9R7I1 A0A1Q3FWP3 T1E210 A0A084VPY1 A0A2Y9D0S9 Q7Q6Q3 A0A2M3YZN6 A0A2M4A6R6 A0A2M4CTE2 A0A0C9RAV3 A0A1B6MTZ5 A0A0A9Y8F5 A0A1B6MJS1 A0A0A9W940 D6WH49 A0A139WK61 A0A2A3EIF9 A0A0K8SJ70 A0A146LJT8 A0A1J1ITG9 A0A0K8TR83 U5EUK7 J9JWI5 A0A2S2N6T1 A0A1W4WFH7 A0A2H8TE41 A0A1W4WF71 A0A1Y1L9M3 T1D3Y2 A0A1B6LLF0 A0A146LWS3 A0A1B6LNE6 A0A2M4A6Y6 A0A2S2QD51 A0A1L8DWX7 B0WMQ5 A0A195BKC8 A0A1L8DKM5 V5I9C0 A0A182Q7T8 A0A067R6P1 A0A1I8MY59 A0A1W4W041 A0A1W4VNV0 A0A3B0K3J1 A0A0J9RN84 Q7KM05 Q7KM08 Q94888 A0A3B0KPW9 A0A3B0KL04 A0A3B0JYW5 A0A1I8MY72 A0A1I8MY64 C8VUZ5 B4PH45 A0A023F4Q8 A0A1I8PTX4 A0A0J9RNB4 Q7KM03 A0A1W4W0Y2 B4LDI5 C9QP46 B3NBX7 A0A3B0K1Q0 M9PE01 A0A1W4W0X8 B4QPK1 B4KYC0 A0A3B0KTL8 B4N572 B4HTS8 Q9XTK9 A0A0J9RNS1 Q7KM04
Pubmed
28756777
22118469
26354079
19121390
21347285
21719571
+ More
20798317 24508170 30249741 26999592 24945155 21282665 24330624 24438588 12364791 25401762 26823975 18362917 19820115 26369729 28004739 24845553 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 25474469
20798317 24508170 30249741 26999592 24945155 21282665 24330624 24438588 12364791 25401762 26823975 18362917 19820115 26369729 28004739 24845553 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 25474469
EMBL
KZ150225
PZC72061.1
AGBW02014596
OWR41361.1
KQ459590
KPI97228.1
+ More
BABH01012265 BABH01012266 KQ461161 KPJ08023.1 ADTU01009957 KQ982686 KYQ52314.1 KQ434938 KZC12139.1 GL888070 EGI68004.1 KQ980734 KYN13784.1 GL437023 EFN71264.1 KQ981305 KYN42921.1 KQ761738 OAD57134.1 KK107871 QOIP01000007 EZA47370.1 RLU20058.1 GDUN01000141 JAN95778.1 KQ414669 KOC64509.1 GAPW01001178 GEHC01000719 JAC12420.1 JAV46926.1 GL766762 EFZ14633.1 GL446286 EFN88086.1 UFQS01000899 UFQT01000899 SSX07557.1 SSX27897.1 KQ435732 KOX77454.1 KQ977276 KYN04455.1 GBYB01003980 JAG73747.1 GFDL01003004 JAV32041.1 GALA01001169 JAA93683.1 ATLV01015071 ATLV01015072 ATLV01015073 ATLV01015074 ATLV01015075 ATLV01015076 ATLV01015077 ATLV01015078 ATLV01015079 KE525001 KFB40025.1 APCN01001498 APCN01001499 AAAB01008960 EAA11623.4 GGFM01000976 MBW21727.1 GGFK01003166 MBW36487.1 GGFL01004419 MBW68597.1 GBYB01003981 JAG73748.1 GEBQ01000563 JAT39414.1 GBHO01014257 GDHC01013022 JAG29347.1 JAQ05607.1 GEBQ01003826 JAT36151.1 GBHO01039673 GBRD01007772 GDHC01007792 JAG03931.1 JAG58049.1 JAQ10837.1 KQ971330 EEZ99754.2 KYB28322.1 KZ288230 PBC31583.1 GBRD01012991 JAG52835.1 GDHC01010266 JAQ08363.1 CVRI01000059 CRL03408.1 GDAI01000714 JAI16889.1 GANO01001428 JAB58443.1 ABLF02040434 ABLF02040435 GGMR01000304 MBY12923.1 GFXV01000572 MBW12377.1 GEZM01061560 JAV70369.1 GALA01001173 JAA93679.1 GEBQ01015491 JAT24486.1 GDHC01006515 JAQ12114.1 GEBQ01014841 JAT25136.1 GGFK01003245 MBW36566.1 GGMS01006461 MBY75664.1 GFDF01003175 JAV10909.1 DS232000 EDS31140.1 KQ976455 KYM85103.1 GFDF01007159 JAV06925.1 GALX01003200 JAB65266.1 AXCN02000881 KK852851 KDR15062.1 OUUW01000012 SPP87242.1 CM002912 KMY97377.1 KMY97381.1 AF152402 AE014296 BT010064 BT022123 AAD45513.1 AAF47781.2 AAG22238.2 AAG22239.2 AAQ22533.1 AAY51518.1 ABI31237.1 AF152399 AAD45510.1 ABI31232.1 ADV37479.1 X99512 CAA67862.1 SPP87241.1 SPP87239.1 SPP87244.1 BT099741 ACV53875.1 CM000159 EDW93282.2 GBBI01002271 JAC16441.1 KMY97378.1 KMY97379.1 KMY97380.1 AF152401 AF152404 AF152405 AF152406 AAD45512.1 AAD45515.1 AAD45516.1 AAD45517.1 ABI31234.1 ABI31235.1 ABI31236.1 ACZ94612.1 CH940647 EDW68923.2 BT099928 ACX36504.1 CH954178 EDV50794.2 SPP87243.1 AGB94068.1 CM000363 EDX09094.1 CH933809 EDW17699.2 SPP87238.1 CH964101 EDW79511.1 CH480817 EDW50349.1 AF152398 AF152400 AAD45509.1 AAD45511.1 AAN11568.1 AAN11569.1 KMY97382.1 AF152403 AAD45514.1 ABI31233.1
BABH01012265 BABH01012266 KQ461161 KPJ08023.1 ADTU01009957 KQ982686 KYQ52314.1 KQ434938 KZC12139.1 GL888070 EGI68004.1 KQ980734 KYN13784.1 GL437023 EFN71264.1 KQ981305 KYN42921.1 KQ761738 OAD57134.1 KK107871 QOIP01000007 EZA47370.1 RLU20058.1 GDUN01000141 JAN95778.1 KQ414669 KOC64509.1 GAPW01001178 GEHC01000719 JAC12420.1 JAV46926.1 GL766762 EFZ14633.1 GL446286 EFN88086.1 UFQS01000899 UFQT01000899 SSX07557.1 SSX27897.1 KQ435732 KOX77454.1 KQ977276 KYN04455.1 GBYB01003980 JAG73747.1 GFDL01003004 JAV32041.1 GALA01001169 JAA93683.1 ATLV01015071 ATLV01015072 ATLV01015073 ATLV01015074 ATLV01015075 ATLV01015076 ATLV01015077 ATLV01015078 ATLV01015079 KE525001 KFB40025.1 APCN01001498 APCN01001499 AAAB01008960 EAA11623.4 GGFM01000976 MBW21727.1 GGFK01003166 MBW36487.1 GGFL01004419 MBW68597.1 GBYB01003981 JAG73748.1 GEBQ01000563 JAT39414.1 GBHO01014257 GDHC01013022 JAG29347.1 JAQ05607.1 GEBQ01003826 JAT36151.1 GBHO01039673 GBRD01007772 GDHC01007792 JAG03931.1 JAG58049.1 JAQ10837.1 KQ971330 EEZ99754.2 KYB28322.1 KZ288230 PBC31583.1 GBRD01012991 JAG52835.1 GDHC01010266 JAQ08363.1 CVRI01000059 CRL03408.1 GDAI01000714 JAI16889.1 GANO01001428 JAB58443.1 ABLF02040434 ABLF02040435 GGMR01000304 MBY12923.1 GFXV01000572 MBW12377.1 GEZM01061560 JAV70369.1 GALA01001173 JAA93679.1 GEBQ01015491 JAT24486.1 GDHC01006515 JAQ12114.1 GEBQ01014841 JAT25136.1 GGFK01003245 MBW36566.1 GGMS01006461 MBY75664.1 GFDF01003175 JAV10909.1 DS232000 EDS31140.1 KQ976455 KYM85103.1 GFDF01007159 JAV06925.1 GALX01003200 JAB65266.1 AXCN02000881 KK852851 KDR15062.1 OUUW01000012 SPP87242.1 CM002912 KMY97377.1 KMY97381.1 AF152402 AE014296 BT010064 BT022123 AAD45513.1 AAF47781.2 AAG22238.2 AAG22239.2 AAQ22533.1 AAY51518.1 ABI31237.1 AF152399 AAD45510.1 ABI31232.1 ADV37479.1 X99512 CAA67862.1 SPP87241.1 SPP87239.1 SPP87244.1 BT099741 ACV53875.1 CM000159 EDW93282.2 GBBI01002271 JAC16441.1 KMY97378.1 KMY97379.1 KMY97380.1 AF152401 AF152404 AF152405 AF152406 AAD45512.1 AAD45515.1 AAD45516.1 AAD45517.1 ABI31234.1 ABI31235.1 ABI31236.1 ACZ94612.1 CH940647 EDW68923.2 BT099928 ACX36504.1 CH954178 EDV50794.2 SPP87243.1 AGB94068.1 CM000363 EDX09094.1 CH933809 EDW17699.2 SPP87238.1 CH964101 EDW79511.1 CH480817 EDW50349.1 AF152398 AF152400 AAD45509.1 AAD45511.1 AAN11568.1 AAN11569.1 KMY97382.1 AF152403 AAD45514.1 ABI31233.1
Proteomes
UP000007151
UP000053268
UP000005204
UP000053240
UP000005205
UP000005203
+ More
UP000075809 UP000076502 UP000007755 UP000078492 UP000000311 UP000078541 UP000053097 UP000279307 UP000053825 UP000008237 UP000053105 UP000078542 UP000030765 UP000075840 UP000007062 UP000007266 UP000242457 UP000183832 UP000007819 UP000192223 UP000002320 UP000078540 UP000075886 UP000027135 UP000095301 UP000192221 UP000268350 UP000000803 UP000002282 UP000095300 UP000008792 UP000008711 UP000000304 UP000009192 UP000007798 UP000001292
UP000075809 UP000076502 UP000007755 UP000078492 UP000000311 UP000078541 UP000053097 UP000279307 UP000053825 UP000008237 UP000053105 UP000078542 UP000030765 UP000075840 UP000007062 UP000007266 UP000242457 UP000183832 UP000007819 UP000192223 UP000002320 UP000078540 UP000075886 UP000027135 UP000095301 UP000192221 UP000268350 UP000000803 UP000002282 UP000095300 UP000008792 UP000008711 UP000000304 UP000009192 UP000007798 UP000001292
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
A0A2W1BCC2
A0A212EIR5
A0A194PW53
H9JQP8
A0A0N1I517
A0A158NA41
+ More
A0A087ZZD4 A0A151WWM6 A0A154PJU9 F4WC24 A0A195DMQ6 E2A5M1 A0A195FR63 A0A310SG71 A0A026VUM0 A0A0P6IXS9 A0A0L7R0Y2 A0A023ESY4 E9IXM1 E2B884 A0A158NA40 A0A336KT93 A0A0N0BIA3 A0A158NA39 A0A195CUX5 A0A0C9R7I1 A0A1Q3FWP3 T1E210 A0A084VPY1 A0A2Y9D0S9 Q7Q6Q3 A0A2M3YZN6 A0A2M4A6R6 A0A2M4CTE2 A0A0C9RAV3 A0A1B6MTZ5 A0A0A9Y8F5 A0A1B6MJS1 A0A0A9W940 D6WH49 A0A139WK61 A0A2A3EIF9 A0A0K8SJ70 A0A146LJT8 A0A1J1ITG9 A0A0K8TR83 U5EUK7 J9JWI5 A0A2S2N6T1 A0A1W4WFH7 A0A2H8TE41 A0A1W4WF71 A0A1Y1L9M3 T1D3Y2 A0A1B6LLF0 A0A146LWS3 A0A1B6LNE6 A0A2M4A6Y6 A0A2S2QD51 A0A1L8DWX7 B0WMQ5 A0A195BKC8 A0A1L8DKM5 V5I9C0 A0A182Q7T8 A0A067R6P1 A0A1I8MY59 A0A1W4W041 A0A1W4VNV0 A0A3B0K3J1 A0A0J9RN84 Q7KM05 Q7KM08 Q94888 A0A3B0KPW9 A0A3B0KL04 A0A3B0JYW5 A0A1I8MY72 A0A1I8MY64 C8VUZ5 B4PH45 A0A023F4Q8 A0A1I8PTX4 A0A0J9RNB4 Q7KM03 A0A1W4W0Y2 B4LDI5 C9QP46 B3NBX7 A0A3B0K1Q0 M9PE01 A0A1W4W0X8 B4QPK1 B4KYC0 A0A3B0KTL8 B4N572 B4HTS8 Q9XTK9 A0A0J9RNS1 Q7KM04
A0A087ZZD4 A0A151WWM6 A0A154PJU9 F4WC24 A0A195DMQ6 E2A5M1 A0A195FR63 A0A310SG71 A0A026VUM0 A0A0P6IXS9 A0A0L7R0Y2 A0A023ESY4 E9IXM1 E2B884 A0A158NA40 A0A336KT93 A0A0N0BIA3 A0A158NA39 A0A195CUX5 A0A0C9R7I1 A0A1Q3FWP3 T1E210 A0A084VPY1 A0A2Y9D0S9 Q7Q6Q3 A0A2M3YZN6 A0A2M4A6R6 A0A2M4CTE2 A0A0C9RAV3 A0A1B6MTZ5 A0A0A9Y8F5 A0A1B6MJS1 A0A0A9W940 D6WH49 A0A139WK61 A0A2A3EIF9 A0A0K8SJ70 A0A146LJT8 A0A1J1ITG9 A0A0K8TR83 U5EUK7 J9JWI5 A0A2S2N6T1 A0A1W4WFH7 A0A2H8TE41 A0A1W4WF71 A0A1Y1L9M3 T1D3Y2 A0A1B6LLF0 A0A146LWS3 A0A1B6LNE6 A0A2M4A6Y6 A0A2S2QD51 A0A1L8DWX7 B0WMQ5 A0A195BKC8 A0A1L8DKM5 V5I9C0 A0A182Q7T8 A0A067R6P1 A0A1I8MY59 A0A1W4W041 A0A1W4VNV0 A0A3B0K3J1 A0A0J9RN84 Q7KM05 Q7KM08 Q94888 A0A3B0KPW9 A0A3B0KL04 A0A3B0JYW5 A0A1I8MY72 A0A1I8MY64 C8VUZ5 B4PH45 A0A023F4Q8 A0A1I8PTX4 A0A0J9RNB4 Q7KM03 A0A1W4W0Y2 B4LDI5 C9QP46 B3NBX7 A0A3B0K1Q0 M9PE01 A0A1W4W0X8 B4QPK1 B4KYC0 A0A3B0KTL8 B4N572 B4HTS8 Q9XTK9 A0A0J9RNS1 Q7KM04
PDB
5G6V
E-value=6.47667e-103,
Score=956
Ontologies
KEGG
GO
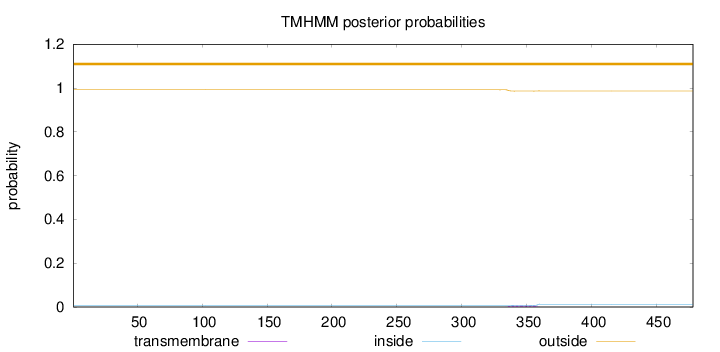
Topology
Length:
478
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.14429
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00812
outside
1 - 478
Population Genetic Test Statistics
Pi
237.912033
Theta
191.144653
Tajima's D
0.863188
CLR
0.099209
CSRT
0.616269186540673
Interpretation
Uncertain
