Gene
KWMTBOMO06487
Pre Gene Modal
BGIBMGA011856
Annotation
putative_ankyrin_repeat_domain_54_[Danaus_plexippus]
Full name
Ankyrin repeat domain-containing protein 54
Alternative Name
Lyn-interacting ankyrin repeat protein
Location in the cell
Nuclear Reliability : 3.284
Sequence
CDS
ATGACAGACTCTGGAGTCGACACTGGAAATGAGAGCAGCGATAATCCTGTGTTTGATCATTCGAAATGTGTTTTGGAATTTAATCTTCCAGCAATTCCTATAACCCAATCTGGTCAGCCAATGCCAGTAGAGTTTTTAGATGAGCCACATGATCATGTCGGAAAAATTAAATGCTCCACAAAAGCAAGACACTGCCGTCTTAAGCATAGATACGGTGGAACAGTTTGCGCATCCAGGAATCACAAGTTGCGGTTCGCGGCTTCCACGAATAACACAGAACTCGTTGAACGATTATTAAAAGCTGGAGCGGATCCTAATTGTTCAGATGAGCACAAAAGAAGCCCATTGCATTTAGCAGCTTGCCGTGGCTATGTCGATGTAGTGAGAATACTTCTGCGTCACGGTGCTAACCCTAACAATAAAGACACACTTGGAAACACACCTCTTCATTTGGCTGCATGTACTAATCACATAACAGTTGTCATTGAACTATTAGATGCTGGAACTGATGTCAGTTCACATGACAGAAATGGCCGCAATCCCATACAATTAGCACAAAGTAAATTAAAGATAATACAAATGCGTCCCAGTGGTGTGGGGCATTTTGAAGTGACTCGTCAATTGATTAGTGAAATTTGCCAAGTTGTAGAAATGATGCTGAAATATATGAAAATGCAGAAGGCTGATGCAGGTGAGCTGGAATCTTTGCGACTGAGGTTGGAGCAAGTGAGTACACGGGAACAAGTTGACTCAGAAGTACAAAATCTACTCGACAGCCTGGACTCACTGAAACTTAGATAA
Protein
MTDSGVDTGNESSDNPVFDHSKCVLEFNLPAIPITQSGQPMPVEFLDEPHDHVGKIKCSTKARHCRLKHRYGGTVCASRNHKLRFAASTNNTELVERLLKAGADPNCSDEHKRSPLHLAACRGYVDVVRILLRHGANPNNKDTLGNTPLHLAACTNHITVVIELLDAGTDVSSHDRNGRNPIQLAQSKLKIIQMRPSGVGHFEVTRQLISEICQVVEMMLKYMKMQKADAGELESLRLRLEQVSTREQVDSEVQNLLDSLDSLKLR
Summary
Description
Plays an important role in regulating intracellular signaling events associated with erythroid terminal differentiation.
Subunit
Interacts (via ankyrin repeat region) with LYN (via SH3-domain) in an activation-independent status of LYN. Forms a multiprotein complex with LYN and HCLS1. Interacts with TSN2, VAV1, DBNL AND LASP1.
Keywords
Acetylation
Alternative splicing
ANK repeat
Complete proteome
Cytoplasm
Nucleus
Phosphoprotein
Reference proteome
Repeat
Feature
chain Ankyrin repeat domain-containing protein 54
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JQQ0
A0A2W1BE37
A0A2A4JMH1
S4P148
A0A2H1VCY2
A0A0L7LUT4
+ More
A0A0N1PGN8 A0A212FN38 I4DKF2 A0A194PUZ1 A0A2S2PR59 A0A2S2Q8X3 J9JY15 A0A1I8NGK4 A0A1I8NQP8 T1PKJ3 A0A2H8THK6 A0A067QZM4 A0A226EFY6 A0A1A9WG43 A0A1A9Z343 A0A1E1XEP4 A0A232EQ91 A0A0A9WBK5 A0A0K8VRJ6 W8BUF2 A0A034WAU2 A0A1S3IDJ6 A0A0A1WVQ3 A0A224XXE3 K7IX54 A0A1D2N5H5 A0A1A9VAT3 A0A1B0B943 A0A0M4F040 B4QNQ3 B3NGM9 A0A0J9RSZ6 B4HLQ6 Q9VT59 A0A224YT69 A0A1W4UMJ5 B4PDX3 A0A131XN07 A0A2T7PWQ9 A0A1B0FGK9 B4J225 B4LCC3 A0A088AH50 B4L923 A0A131YZY5 B3MA58 A0A2A3ETS7 V9L5C1 A0A1Z5LCI9 A0A3Q7Q7L9 B4MN14 L5M9H6 A0A2Y9GVR3 K9IX63 A0A2U3YBB5 S7NLD8 A0A2U3WP12 A0A1B6F4M5 A0A293MR29 G7N3T6 G7PFE1 A0A1A8REN4 A0A1A8QLV6 U6D2K4 E2AN12 A0A3Q0T2T6 M1EEA7 H9H799 H9GMB2 A0A195BSK0 A0A195DI55 H9FA25 G1PPQ7 E9J423 A0A384DA99 L5KI94 A0A2J8UWC2 K7C930 Q6NXT1 A0A087TKK3 A0A2K5VZQ7 A0A0D9R3L9 A0A2K5M662 I2CWG3 A0A2Y9KM81 W5PY71 A0A341CST1 A0A2K6NBW8 A0A3P4KHP0 M3YNA7 E2QSJ6 A0A3Q7VC82
A0A0N1PGN8 A0A212FN38 I4DKF2 A0A194PUZ1 A0A2S2PR59 A0A2S2Q8X3 J9JY15 A0A1I8NGK4 A0A1I8NQP8 T1PKJ3 A0A2H8THK6 A0A067QZM4 A0A226EFY6 A0A1A9WG43 A0A1A9Z343 A0A1E1XEP4 A0A232EQ91 A0A0A9WBK5 A0A0K8VRJ6 W8BUF2 A0A034WAU2 A0A1S3IDJ6 A0A0A1WVQ3 A0A224XXE3 K7IX54 A0A1D2N5H5 A0A1A9VAT3 A0A1B0B943 A0A0M4F040 B4QNQ3 B3NGM9 A0A0J9RSZ6 B4HLQ6 Q9VT59 A0A224YT69 A0A1W4UMJ5 B4PDX3 A0A131XN07 A0A2T7PWQ9 A0A1B0FGK9 B4J225 B4LCC3 A0A088AH50 B4L923 A0A131YZY5 B3MA58 A0A2A3ETS7 V9L5C1 A0A1Z5LCI9 A0A3Q7Q7L9 B4MN14 L5M9H6 A0A2Y9GVR3 K9IX63 A0A2U3YBB5 S7NLD8 A0A2U3WP12 A0A1B6F4M5 A0A293MR29 G7N3T6 G7PFE1 A0A1A8REN4 A0A1A8QLV6 U6D2K4 E2AN12 A0A3Q0T2T6 M1EEA7 H9H799 H9GMB2 A0A195BSK0 A0A195DI55 H9FA25 G1PPQ7 E9J423 A0A384DA99 L5KI94 A0A2J8UWC2 K7C930 Q6NXT1 A0A087TKK3 A0A2K5VZQ7 A0A0D9R3L9 A0A2K5M662 I2CWG3 A0A2Y9KM81 W5PY71 A0A341CST1 A0A2K6NBW8 A0A3P4KHP0 M3YNA7 E2QSJ6 A0A3Q7VC82
Pubmed
19121390
28756777
23622113
26227816
26354079
22118469
+ More
22651552 25315136 24845553 28503490 28648823 25401762 26823975 24495485 25348373 25830018 20075255 27289101 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28797301 17550304 28049606 18057021 26830274 24402279 28528879 22002653 20798317 23236062 17495919 21881562 25319552 21993624 21282665 23258410 15164055 16136131 15461802 14702039 10591208 15489334 18691976 19413330 19369195 20068231 21406692 24275569 20809919 25362486 16341006
22651552 25315136 24845553 28503490 28648823 25401762 26823975 24495485 25348373 25830018 20075255 27289101 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28797301 17550304 28049606 18057021 26830274 24402279 28528879 22002653 20798317 23236062 17495919 21881562 25319552 21993624 21282665 23258410 15164055 16136131 15461802 14702039 10591208 15489334 18691976 19413330 19369195 20068231 21406692 24275569 20809919 25362486 16341006
EMBL
BABH01012275
KZ150297
PZC71557.1
NWSH01000980
PCG73275.1
GAIX01009061
+ More
JAA83499.1 ODYU01001870 SOQ38675.1 JTDY01000052 KOB79154.1 KQ461161 KPJ08020.1 AGBW02007611 OWR55148.1 AK401770 BAM18392.1 KQ459590 KPI97226.1 GGMR01019179 MBY31798.1 GGMS01004817 MBY74020.1 ABLF02030504 KA648615 AFP63244.1 GFXV01001705 MBW13510.1 KK852854 KDR14976.1 LNIX01000004 OXA55721.1 GFAC01001451 JAT97737.1 NNAY01002826 OXU20487.1 GBHO01041369 GBHO01041368 GBHO01041367 GBHO01041366 GBHO01041365 GBHO01034967 GDHC01019025 GDHC01013409 JAG02235.1 JAG02236.1 JAG02237.1 JAG02238.1 JAG02239.1 JAG08637.1 JAP99603.1 JAQ05220.1 GDHF01010812 JAI41502.1 GAMC01013801 JAB92754.1 GAKP01008030 GAKP01008029 JAC50923.1 GBXI01011692 JAD02600.1 GFTR01003236 JAW13190.1 AAZX01008221 LJIJ01000206 ODN00472.1 JXJN01010264 CP012525 ALC44082.1 CM000363 EDX09920.1 CH954178 EDV51265.1 CM002912 KMY98782.1 CH480815 EDW40938.1 AE014296 BT023914 AAF50195.1 ABA81848.1 GFPF01006038 MAA17184.1 CM000159 EDW93969.1 GEFH01001071 JAP67510.1 PZQS01000001 PVD37871.1 CCAG010013633 CH916366 EDV95950.1 CH940647 EDW68768.1 KRF84010.1 CH933816 EDW17198.1 GEDV01004455 JAP84102.1 CH902618 EDV39072.1 KZ288189 PBC34682.1 JW874681 AFP07198.1 GFJQ02002236 JAW04734.1 CH963847 EDW73570.1 KB102787 ELK35001.1 GABZ01007204 JAA46321.1 KE164518 EPQ18306.1 GECZ01024645 JAS45124.1 GFWV01018596 MAA43324.1 CM001262 EHH20218.1 CM001285 EHH65825.1 HAEH01003551 HAEI01008241 SBS04496.1 HAEF01008300 HAEG01013116 SBR94099.1 HAAF01004293 CCP76118.1 GL441034 EFN65170.1 JP005631 AER94228.1 AAWZ02028380 KQ976419 KYM89275.1 KQ980824 KYN12517.1 JU327728 AFE71484.1 AAPE02044881 GL768069 EFZ12435.1 KB030755 ELK10243.1 NDHI03003442 PNJ49547.1 AACZ04068050 GABC01010058 GABE01005663 NBAG03000283 JAA01280.1 JAA39076.1 PNI49000.1 CR456471 AK127583 Z97630 BC014641 BC066909 KK115653 KFM65642.1 AQIA01007032 AQIB01136613 AQIB01136614 AQIB01136615 AQIB01136616 AQIB01136617 JV635958 AFJ71298.1 AMGL01083896 AMGL01083897 CYRY02000473 VCW50000.1 AEYP01012683 AAEX03007325
JAA83499.1 ODYU01001870 SOQ38675.1 JTDY01000052 KOB79154.1 KQ461161 KPJ08020.1 AGBW02007611 OWR55148.1 AK401770 BAM18392.1 KQ459590 KPI97226.1 GGMR01019179 MBY31798.1 GGMS01004817 MBY74020.1 ABLF02030504 KA648615 AFP63244.1 GFXV01001705 MBW13510.1 KK852854 KDR14976.1 LNIX01000004 OXA55721.1 GFAC01001451 JAT97737.1 NNAY01002826 OXU20487.1 GBHO01041369 GBHO01041368 GBHO01041367 GBHO01041366 GBHO01041365 GBHO01034967 GDHC01019025 GDHC01013409 JAG02235.1 JAG02236.1 JAG02237.1 JAG02238.1 JAG02239.1 JAG08637.1 JAP99603.1 JAQ05220.1 GDHF01010812 JAI41502.1 GAMC01013801 JAB92754.1 GAKP01008030 GAKP01008029 JAC50923.1 GBXI01011692 JAD02600.1 GFTR01003236 JAW13190.1 AAZX01008221 LJIJ01000206 ODN00472.1 JXJN01010264 CP012525 ALC44082.1 CM000363 EDX09920.1 CH954178 EDV51265.1 CM002912 KMY98782.1 CH480815 EDW40938.1 AE014296 BT023914 AAF50195.1 ABA81848.1 GFPF01006038 MAA17184.1 CM000159 EDW93969.1 GEFH01001071 JAP67510.1 PZQS01000001 PVD37871.1 CCAG010013633 CH916366 EDV95950.1 CH940647 EDW68768.1 KRF84010.1 CH933816 EDW17198.1 GEDV01004455 JAP84102.1 CH902618 EDV39072.1 KZ288189 PBC34682.1 JW874681 AFP07198.1 GFJQ02002236 JAW04734.1 CH963847 EDW73570.1 KB102787 ELK35001.1 GABZ01007204 JAA46321.1 KE164518 EPQ18306.1 GECZ01024645 JAS45124.1 GFWV01018596 MAA43324.1 CM001262 EHH20218.1 CM001285 EHH65825.1 HAEH01003551 HAEI01008241 SBS04496.1 HAEF01008300 HAEG01013116 SBR94099.1 HAAF01004293 CCP76118.1 GL441034 EFN65170.1 JP005631 AER94228.1 AAWZ02028380 KQ976419 KYM89275.1 KQ980824 KYN12517.1 JU327728 AFE71484.1 AAPE02044881 GL768069 EFZ12435.1 KB030755 ELK10243.1 NDHI03003442 PNJ49547.1 AACZ04068050 GABC01010058 GABE01005663 NBAG03000283 JAA01280.1 JAA39076.1 PNI49000.1 CR456471 AK127583 Z97630 BC014641 BC066909 KK115653 KFM65642.1 AQIA01007032 AQIB01136613 AQIB01136614 AQIB01136615 AQIB01136616 AQIB01136617 JV635958 AFJ71298.1 AMGL01083896 AMGL01083897 CYRY02000473 VCW50000.1 AEYP01012683 AAEX03007325
Proteomes
UP000005204
UP000218220
UP000037510
UP000053240
UP000007151
UP000053268
+ More
UP000007819 UP000095301 UP000095300 UP000027135 UP000198287 UP000091820 UP000092445 UP000215335 UP000085678 UP000002358 UP000094527 UP000078200 UP000092460 UP000092553 UP000000304 UP000008711 UP000001292 UP000000803 UP000192221 UP000002282 UP000245119 UP000092444 UP000001070 UP000008792 UP000005203 UP000009192 UP000007801 UP000242457 UP000286641 UP000007798 UP000248481 UP000245341 UP000245340 UP000009130 UP000000311 UP000261340 UP000002280 UP000001646 UP000078540 UP000078492 UP000001074 UP000261680 UP000010552 UP000002277 UP000005640 UP000054359 UP000233100 UP000029965 UP000233060 UP000248482 UP000002356 UP000252040 UP000233200 UP000000715 UP000002254 UP000286642
UP000007819 UP000095301 UP000095300 UP000027135 UP000198287 UP000091820 UP000092445 UP000215335 UP000085678 UP000002358 UP000094527 UP000078200 UP000092460 UP000092553 UP000000304 UP000008711 UP000001292 UP000000803 UP000192221 UP000002282 UP000245119 UP000092444 UP000001070 UP000008792 UP000005203 UP000009192 UP000007801 UP000242457 UP000286641 UP000007798 UP000248481 UP000245341 UP000245340 UP000009130 UP000000311 UP000261340 UP000002280 UP000001646 UP000078540 UP000078492 UP000001074 UP000261680 UP000010552 UP000002277 UP000005640 UP000054359 UP000233100 UP000029965 UP000233060 UP000248482 UP000002356 UP000252040 UP000233200 UP000000715 UP000002254 UP000286642
Interpro
SUPFAM
SSF48403
SSF48403
Gene 3D
CDD
ProteinModelPortal
H9JQQ0
A0A2W1BE37
A0A2A4JMH1
S4P148
A0A2H1VCY2
A0A0L7LUT4
+ More
A0A0N1PGN8 A0A212FN38 I4DKF2 A0A194PUZ1 A0A2S2PR59 A0A2S2Q8X3 J9JY15 A0A1I8NGK4 A0A1I8NQP8 T1PKJ3 A0A2H8THK6 A0A067QZM4 A0A226EFY6 A0A1A9WG43 A0A1A9Z343 A0A1E1XEP4 A0A232EQ91 A0A0A9WBK5 A0A0K8VRJ6 W8BUF2 A0A034WAU2 A0A1S3IDJ6 A0A0A1WVQ3 A0A224XXE3 K7IX54 A0A1D2N5H5 A0A1A9VAT3 A0A1B0B943 A0A0M4F040 B4QNQ3 B3NGM9 A0A0J9RSZ6 B4HLQ6 Q9VT59 A0A224YT69 A0A1W4UMJ5 B4PDX3 A0A131XN07 A0A2T7PWQ9 A0A1B0FGK9 B4J225 B4LCC3 A0A088AH50 B4L923 A0A131YZY5 B3MA58 A0A2A3ETS7 V9L5C1 A0A1Z5LCI9 A0A3Q7Q7L9 B4MN14 L5M9H6 A0A2Y9GVR3 K9IX63 A0A2U3YBB5 S7NLD8 A0A2U3WP12 A0A1B6F4M5 A0A293MR29 G7N3T6 G7PFE1 A0A1A8REN4 A0A1A8QLV6 U6D2K4 E2AN12 A0A3Q0T2T6 M1EEA7 H9H799 H9GMB2 A0A195BSK0 A0A195DI55 H9FA25 G1PPQ7 E9J423 A0A384DA99 L5KI94 A0A2J8UWC2 K7C930 Q6NXT1 A0A087TKK3 A0A2K5VZQ7 A0A0D9R3L9 A0A2K5M662 I2CWG3 A0A2Y9KM81 W5PY71 A0A341CST1 A0A2K6NBW8 A0A3P4KHP0 M3YNA7 E2QSJ6 A0A3Q7VC82
A0A0N1PGN8 A0A212FN38 I4DKF2 A0A194PUZ1 A0A2S2PR59 A0A2S2Q8X3 J9JY15 A0A1I8NGK4 A0A1I8NQP8 T1PKJ3 A0A2H8THK6 A0A067QZM4 A0A226EFY6 A0A1A9WG43 A0A1A9Z343 A0A1E1XEP4 A0A232EQ91 A0A0A9WBK5 A0A0K8VRJ6 W8BUF2 A0A034WAU2 A0A1S3IDJ6 A0A0A1WVQ3 A0A224XXE3 K7IX54 A0A1D2N5H5 A0A1A9VAT3 A0A1B0B943 A0A0M4F040 B4QNQ3 B3NGM9 A0A0J9RSZ6 B4HLQ6 Q9VT59 A0A224YT69 A0A1W4UMJ5 B4PDX3 A0A131XN07 A0A2T7PWQ9 A0A1B0FGK9 B4J225 B4LCC3 A0A088AH50 B4L923 A0A131YZY5 B3MA58 A0A2A3ETS7 V9L5C1 A0A1Z5LCI9 A0A3Q7Q7L9 B4MN14 L5M9H6 A0A2Y9GVR3 K9IX63 A0A2U3YBB5 S7NLD8 A0A2U3WP12 A0A1B6F4M5 A0A293MR29 G7N3T6 G7PFE1 A0A1A8REN4 A0A1A8QLV6 U6D2K4 E2AN12 A0A3Q0T2T6 M1EEA7 H9H799 H9GMB2 A0A195BSK0 A0A195DI55 H9FA25 G1PPQ7 E9J423 A0A384DA99 L5KI94 A0A2J8UWC2 K7C930 Q6NXT1 A0A087TKK3 A0A2K5VZQ7 A0A0D9R3L9 A0A2K5M662 I2CWG3 A0A2Y9KM81 W5PY71 A0A341CST1 A0A2K6NBW8 A0A3P4KHP0 M3YNA7 E2QSJ6 A0A3Q7VC82
PDB
1N0R
E-value=4.01532e-22,
Score=257
Ontologies
GO
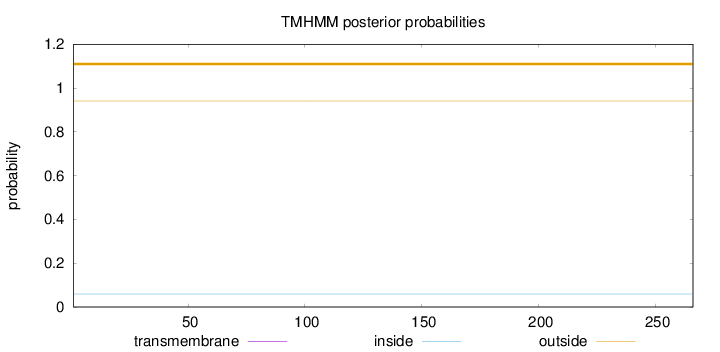
Topology
Subcellular location
Nucleus
Shuttles between nucleus and cytoplasm during the cell cycle. EPO stimulation induces nuclear accumulation (By similarity). With evidence from 7 publications.
Cytoplasm Shuttles between nucleus and cytoplasm during the cell cycle. EPO stimulation induces nuclear accumulation (By similarity). With evidence from 7 publications.
Midbody Shuttles between nucleus and cytoplasm during the cell cycle. EPO stimulation induces nuclear accumulation (By similarity). With evidence from 7 publications.
Cytoplasm Shuttles between nucleus and cytoplasm during the cell cycle. EPO stimulation induces nuclear accumulation (By similarity). With evidence from 7 publications.
Midbody Shuttles between nucleus and cytoplasm during the cell cycle. EPO stimulation induces nuclear accumulation (By similarity). With evidence from 7 publications.
Length:
266
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00207
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05928
outside
1 - 266
Population Genetic Test Statistics
Pi
179.783713
Theta
236.475835
Tajima's D
-0.755072
CLR
1131.869712
CSRT
0.185690715464227
Interpretation
Uncertain
