Gene
KWMTBOMO06486
Pre Gene Modal
BGIBMGA011622
Annotation
PREDICTED:_sphingosine_kinase_1-like_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 2.368
Sequence
CDS
ATGTTTGGTAAGGACGTGTCGAAAAATATTGAAGGTGACTCGGTTCCGAAAGATCATGTGTATTTGGAAGAAACGTTTTATATACTCTCTAAGAAGAATTCAGTGTTTCGGGTTCGTCTAACATCTAAAGGGCTGTCTTTAACAAAAGAAACTGATGGTAGTTCTAAAGAACAAACTATTTTGTTGTGCGACATTATAGGCAGTAAATGTATGAGAAGCAAAAGACGGCGACACGGAGCTAGCTCTTGTGTCTGCAGTTCGTTGGTGGGTCCTCAACAGCTAAAAGTTGTGGAGGAGAACAGTGGCGATCTGGACGAAAATGACATTAGTGCGTATTTGTACATTTACTCATATATTTTGAAACGAAGCCGTCGCAGTTGGAAGAGGGAAAGAACAACAATAACCCTTCGCTTCAGGTCTTTCGATAAATACGAAGATAATAACAGAGAGGCGCAAAAGTGGCGTACATTTATAAAATGCTTGATAGCCGGTCAGCCGATTACGTATGTGCCACCGCTCAACGATAAGAAAATACTTATACTTCTCAATCCAAAATCCGGACCTGGGAAGGCTAGAGAATTGTTTCAGACCAAGGTGGCTCCGATATTACAAGAAGCGGAAGTGCCGTATGACCTTCATGTCACTAAATATGCTCAATTTGCCCGCGAATTCGTTCGTACGCGTAATGTTTATGCTTGGCGTACTATTGTGGCGGTGGGTGGAGACGGTGTGTTGTTTGAAATCCTTAATGGTATGTTTGAGCGCTTGGATTGGCAACAAGCACTGTCGGAGATTCCTCTCGCCATCATCCCCTGTGGGTCAGGAAATGGATTGGCACGAACTATTTGCCACCTCTACAATGAGCCGTACATCACTCAAAATTTAACCGGCATTTCGATGGCTCTAGTTAAGAACAGAATCACTCCGATGGACGTCGTTCGCATAGAAACAAAAACGAAAATCATGTTTTCCTTTCTATCGGTTGGATGGGGCTTATTGTCAGACATCGACATTGAGAGCGAGAGATTACGAGCAATCGGAGGTCAGAGATTTGCGGTGTGGGCAGTTGCGAGATTGCTCGGACTGAGGCGGTATCGGGGAGTTATCAGCTACGTTAAAATTAAGGATGTCACTAATCTTCCCAAGCCGAAACAACCGTTGGCTCTCAGCCACAGCGTGAGCCAGGATGGGGCACTCGATTCCCCCGACGCGGAAGCGTTTATTGACTGCGATGAAAATTCCGAAGTGTTTAGCAACGCGGTCAACGGGAAGCACCATCAGCGGGTTGATTCGTGGTATTCTGTTAATTCACGGAGAAGCGCATTTTACAGTACGCGTGGTTCGGAATATCATAGCGTCACCAGCGGTGGTTCGGAGATGCGATCGCCGGTACACGCTTGTATGCACGGGCCGGCTTCGCATCTGCCCTCGTTAGTATCTCAGCTACCGTCGCATTGGGTCCACGAAGAAGGAGAGTTTGTCATGGTTCACGTAGCGTATCAGCCGTACATTGGAGAAGATTTCTTGTTTGCACCGCGATCCACGCTCTCCGACGGCGTGATGTGGATGTTGATAATCAAAGCTGGGATTTCTCGATCACAGCTACTCTCATTCCTTCTCGGCATCGGTCAAGGGTCGCATGCGGAAATCAATAATGAATATGTAAAAATGATTCCTGTTAGTGCATTTAGAATAGTACCGGAAGGTTCGAGCGGATATCTCACCGTTGACGGGGAGCTCGTAGAGTACGGACCTATACAAGCAGAGATCTTTCCGAATGTAGTAAATCTCATAGTGCCAGATTTAAAATAA
Protein
MFGKDVSKNIEGDSVPKDHVYLEETFYILSKKNSVFRVRLTSKGLSLTKETDGSSKEQTILLCDIIGSKCMRSKRRRHGASSCVCSSLVGPQQLKVVEENSGDLDENDISAYLYIYSYILKRSRRSWKRERTTITLRFRSFDKYEDNNREAQKWRTFIKCLIAGQPITYVPPLNDKKILILLNPKSGPGKARELFQTKVAPILQEAEVPYDLHVTKYAQFAREFVRTRNVYAWRTIVAVGGDGVLFEILNGMFERLDWQQALSEIPLAIIPCGSGNGLARTICHLYNEPYITQNLTGISMALVKNRITPMDVVRIETKTKIMFSFLSVGWGLLSDIDIESERLRAIGGQRFAVWAVARLLGLRRYRGVISYVKIKDVTNLPKPKQPLALSHSVSQDGALDSPDAEAFIDCDENSEVFSNAVNGKHHQRVDSWYSVNSRRSAFYSTRGSEYHSVTSGGSEMRSPVHACMHGPASHLPSLVSQLPSHWVHEEGEFVMVHVAYQPYIGEDFLFAPRSTLSDGVMWMLIIKAGISRSQLLSFLLGIGQGSHAEINNEYVKMIPVSAFRIVPEGSSGYLTVDGELVEYGPIQAEIFPNVVNLIVPDLK
Summary
Uniprot
H9JQ16
A0A2W1BFE4
A0A2A4JNW5
A0A194Q1F0
A0A0N0PAT2
A0A1Q3G0Z1
+ More
Q16YK7 U5ESN1 A0A1L8DZB5 A0A2C9GPF5 A0A182UZW9 Q7QIP4 A0A182ILN7 A0A182P132 A0A2J7QKZ7 A0A182JTG2 A0A1B0CMB3 A0A2M4BHW4 A0A2M3Z7Q2 W5JA25 A0A154NYJ3 A0A1L8DYS7 A0A0L7RHQ2 A0A310SC29 A0A2A3EUE7 A0A088AGY3 A0A182Q368 D6WDP5 E2AFH3 A0A0C9RFJ1 A0A026WC95 A0A151XAS8 A0A195DHT8 A0A158P1Q5 A0A195EQR8 A0A195BSR1 A0A195CMA1 A0A1Y1MCI8 A0A067R932 E0VL53 A0A1B6HBE2 A0A1B6MNW0 A0A2M4D0Q4 K7IX55 A0A1B6DK62 W8CCJ2 A0A2M3Z8M5 A0A0K8VIY2 N6UM55 A0A0A1XMJ5 A0A034WI04 A0A0P4VQG4 A0A023FBP6 A0A0M4E862 R4WDZ4 B4H402 A0A0A9WQ72 B4KWK9 B3NF21 Q29E36 B4IYW4 B4LEC0 B4PCW2 B3M7F2 A0A0L0CI52 A0A1W4W1V7 A0A3B0J5A4 B4QP12 Q9VZW0 B4N6I1 A0A224XK94 A0A0M9A1I0 A0A1I8N340 A0A1A9XHI0 A0A1B0BHV1 B4HTJ0 A0A336LKV8 A0A1A9VED7 A0A1A9Z4L0 A0A2H8TZI5 J9K841 A0A1B0FA55 A0A1A9W7I7 A0A1B0AIY1 A0A1B0B759 A0A1B0G1S5 A0A1A9Y1D9 A0A0M3QZI6 A0A0L0C859 A0A2S2R5T4 A0A0A1WZY3 A0A0A1WFF0 A0A0A1WJ54 B3NUD3 Q9VYY8
Q16YK7 U5ESN1 A0A1L8DZB5 A0A2C9GPF5 A0A182UZW9 Q7QIP4 A0A182ILN7 A0A182P132 A0A2J7QKZ7 A0A182JTG2 A0A1B0CMB3 A0A2M4BHW4 A0A2M3Z7Q2 W5JA25 A0A154NYJ3 A0A1L8DYS7 A0A0L7RHQ2 A0A310SC29 A0A2A3EUE7 A0A088AGY3 A0A182Q368 D6WDP5 E2AFH3 A0A0C9RFJ1 A0A026WC95 A0A151XAS8 A0A195DHT8 A0A158P1Q5 A0A195EQR8 A0A195BSR1 A0A195CMA1 A0A1Y1MCI8 A0A067R932 E0VL53 A0A1B6HBE2 A0A1B6MNW0 A0A2M4D0Q4 K7IX55 A0A1B6DK62 W8CCJ2 A0A2M3Z8M5 A0A0K8VIY2 N6UM55 A0A0A1XMJ5 A0A034WI04 A0A0P4VQG4 A0A023FBP6 A0A0M4E862 R4WDZ4 B4H402 A0A0A9WQ72 B4KWK9 B3NF21 Q29E36 B4IYW4 B4LEC0 B4PCW2 B3M7F2 A0A0L0CI52 A0A1W4W1V7 A0A3B0J5A4 B4QP12 Q9VZW0 B4N6I1 A0A224XK94 A0A0M9A1I0 A0A1I8N340 A0A1A9XHI0 A0A1B0BHV1 B4HTJ0 A0A336LKV8 A0A1A9VED7 A0A1A9Z4L0 A0A2H8TZI5 J9K841 A0A1B0FA55 A0A1A9W7I7 A0A1B0AIY1 A0A1B0B759 A0A1B0G1S5 A0A1A9Y1D9 A0A0M3QZI6 A0A0L0C859 A0A2S2R5T4 A0A0A1WZY3 A0A0A1WFF0 A0A0A1WJ54 B3NUD3 Q9VYY8
Pubmed
19121390
28756777
26354079
17510324
12364791
20920257
+ More
23761445 18362917 19820115 20798317 24508170 30249741 21347285 28004739 24845553 20566863 20075255 24495485 23537049 25830018 25348373 27129103 25474469 23691247 17994087 25401762 15632085 17550304 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136
23761445 18362917 19820115 20798317 24508170 30249741 21347285 28004739 24845553 20566863 20075255 24495485 23537049 25830018 25348373 27129103 25474469 23691247 17994087 25401762 15632085 17550304 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136
EMBL
BABH01012276
BABH01012277
KZ150297
PZC71556.1
NWSH01000980
PCG73274.1
+ More
KQ459590 KPI97225.1 KQ461161 KPJ08019.1 GFDL01001574 JAV33471.1 CH477515 CH477298 EAT39700.1 EAT44350.1 GANO01003189 JAB56682.1 GFDF01002452 JAV11632.1 APCN01003701 APCN01003702 AAAB01008807 EAA04298.3 NEVH01013255 PNF29259.1 AJWK01018544 AJWK01018545 AJWK01018546 AJWK01018547 AJWK01018548 GGFJ01003257 MBW52398.1 GGFM01003784 MBW24535.1 ADMH02002070 ETN59614.1 KQ434775 KZC04088.1 GFDF01002498 JAV11586.1 KQ414591 KOC70276.1 KQ763877 OAD54655.1 KZ288189 PBC34681.1 AXCN02000985 KQ971322 EFA00810.1 GL439108 EFN67866.1 GBYB01011892 GBYB01011893 GBYB01011894 JAG81659.1 JAG81660.1 JAG81661.1 KK107274 QOIP01000008 EZA53682.1 RLU19214.1 KQ982335 KYQ57482.1 KQ980824 KYN12460.1 ADTU01006749 ADTU01006750 ADTU01006751 KQ982021 KYN30521.1 KQ976419 KYM89335.1 KQ977595 KYN01597.1 GEZM01037634 JAV82015.1 KK852854 KDR14977.1 DS235269 EEB14109.1 GECU01035777 JAS71929.1 GEBQ01002339 JAT37638.1 GGFL01006500 MBW70678.1 AAZX01000586 GEDC01011298 JAS26000.1 GAMC01003601 GAMC01003600 JAC02955.1 GGFM01004101 MBW24852.1 GDHF01019055 GDHF01013794 GDHF01007482 JAI33259.1 JAI38520.1 JAI44832.1 APGK01027154 APGK01027155 KB740648 ENN79792.1 GBXI01005538 GBXI01002071 JAD08754.1 JAD12221.1 GAKP01005192 GAKP01005191 JAC53760.1 GDKW01001353 JAI55242.1 GBBI01000293 JAC18419.1 CP012525 ALC43016.1 AK417989 BAN21204.1 CH479208 EDW31117.1 GBHO01036594 JAG07010.1 CH933809 EDW19638.1 CH954178 EDV50363.2 CH379070 EAL30227.2 CH916366 EDV97672.1 CH940647 EDW70096.1 CM000159 EDW93866.1 CH902618 EDV39850.1 JRES01000364 KNC31920.1 OUUW01000002 SPP77124.1 CM000363 CM002912 EDX09007.1 KMY97225.1 AE014296 AY069417 AAF47706.3 AAL39562.1 CH964161 EDW79970.1 GFTR01007404 JAW09022.1 KQ435794 KOX73983.1 JXJN01014603 CH480817 EDW50261.1 UFQS01000004 UFQT01000004 SSW96866.1 SSX17253.1 GFXV01007869 MBW19674.1 ABLF02030491 ABLF02030504 CCAG010006049 JXJN01009403 CCAG010007360 CP012528 ALC49483.1 JRES01000777 KNC28417.1 GGMS01015519 MBY84722.1 GBXI01009895 JAD04397.1 GBXI01016665 JAC97626.1 GBXI01015435 JAC98856.1 CH954180 EDV46048.1 AE014298 AY119652 AAF48045.1 AAM50306.1 AAN09634.1
KQ459590 KPI97225.1 KQ461161 KPJ08019.1 GFDL01001574 JAV33471.1 CH477515 CH477298 EAT39700.1 EAT44350.1 GANO01003189 JAB56682.1 GFDF01002452 JAV11632.1 APCN01003701 APCN01003702 AAAB01008807 EAA04298.3 NEVH01013255 PNF29259.1 AJWK01018544 AJWK01018545 AJWK01018546 AJWK01018547 AJWK01018548 GGFJ01003257 MBW52398.1 GGFM01003784 MBW24535.1 ADMH02002070 ETN59614.1 KQ434775 KZC04088.1 GFDF01002498 JAV11586.1 KQ414591 KOC70276.1 KQ763877 OAD54655.1 KZ288189 PBC34681.1 AXCN02000985 KQ971322 EFA00810.1 GL439108 EFN67866.1 GBYB01011892 GBYB01011893 GBYB01011894 JAG81659.1 JAG81660.1 JAG81661.1 KK107274 QOIP01000008 EZA53682.1 RLU19214.1 KQ982335 KYQ57482.1 KQ980824 KYN12460.1 ADTU01006749 ADTU01006750 ADTU01006751 KQ982021 KYN30521.1 KQ976419 KYM89335.1 KQ977595 KYN01597.1 GEZM01037634 JAV82015.1 KK852854 KDR14977.1 DS235269 EEB14109.1 GECU01035777 JAS71929.1 GEBQ01002339 JAT37638.1 GGFL01006500 MBW70678.1 AAZX01000586 GEDC01011298 JAS26000.1 GAMC01003601 GAMC01003600 JAC02955.1 GGFM01004101 MBW24852.1 GDHF01019055 GDHF01013794 GDHF01007482 JAI33259.1 JAI38520.1 JAI44832.1 APGK01027154 APGK01027155 KB740648 ENN79792.1 GBXI01005538 GBXI01002071 JAD08754.1 JAD12221.1 GAKP01005192 GAKP01005191 JAC53760.1 GDKW01001353 JAI55242.1 GBBI01000293 JAC18419.1 CP012525 ALC43016.1 AK417989 BAN21204.1 CH479208 EDW31117.1 GBHO01036594 JAG07010.1 CH933809 EDW19638.1 CH954178 EDV50363.2 CH379070 EAL30227.2 CH916366 EDV97672.1 CH940647 EDW70096.1 CM000159 EDW93866.1 CH902618 EDV39850.1 JRES01000364 KNC31920.1 OUUW01000002 SPP77124.1 CM000363 CM002912 EDX09007.1 KMY97225.1 AE014296 AY069417 AAF47706.3 AAL39562.1 CH964161 EDW79970.1 GFTR01007404 JAW09022.1 KQ435794 KOX73983.1 JXJN01014603 CH480817 EDW50261.1 UFQS01000004 UFQT01000004 SSW96866.1 SSX17253.1 GFXV01007869 MBW19674.1 ABLF02030491 ABLF02030504 CCAG010006049 JXJN01009403 CCAG010007360 CP012528 ALC49483.1 JRES01000777 KNC28417.1 GGMS01015519 MBY84722.1 GBXI01009895 JAD04397.1 GBXI01016665 JAC97626.1 GBXI01015435 JAC98856.1 CH954180 EDV46048.1 AE014298 AY119652 AAF48045.1 AAM50306.1 AAN09634.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000008820
UP000075840
+ More
UP000075903 UP000007062 UP000075880 UP000075885 UP000235965 UP000075881 UP000092461 UP000000673 UP000076502 UP000053825 UP000242457 UP000005203 UP000075886 UP000007266 UP000000311 UP000053097 UP000279307 UP000075809 UP000078492 UP000005205 UP000078541 UP000078540 UP000078542 UP000027135 UP000009046 UP000002358 UP000019118 UP000092553 UP000008744 UP000009192 UP000008711 UP000001819 UP000001070 UP000008792 UP000002282 UP000007801 UP000037069 UP000192221 UP000268350 UP000000304 UP000000803 UP000007798 UP000053105 UP000095301 UP000092443 UP000092460 UP000001292 UP000078200 UP000092445 UP000007819 UP000092444 UP000091820
UP000075903 UP000007062 UP000075880 UP000075885 UP000235965 UP000075881 UP000092461 UP000000673 UP000076502 UP000053825 UP000242457 UP000005203 UP000075886 UP000007266 UP000000311 UP000053097 UP000279307 UP000075809 UP000078492 UP000005205 UP000078541 UP000078540 UP000078542 UP000027135 UP000009046 UP000002358 UP000019118 UP000092553 UP000008744 UP000009192 UP000008711 UP000001819 UP000001070 UP000008792 UP000002282 UP000007801 UP000037069 UP000192221 UP000268350 UP000000304 UP000000803 UP000007798 UP000053105 UP000095301 UP000092443 UP000092460 UP000001292 UP000078200 UP000092445 UP000007819 UP000092444 UP000091820
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JQ16
A0A2W1BFE4
A0A2A4JNW5
A0A194Q1F0
A0A0N0PAT2
A0A1Q3G0Z1
+ More
Q16YK7 U5ESN1 A0A1L8DZB5 A0A2C9GPF5 A0A182UZW9 Q7QIP4 A0A182ILN7 A0A182P132 A0A2J7QKZ7 A0A182JTG2 A0A1B0CMB3 A0A2M4BHW4 A0A2M3Z7Q2 W5JA25 A0A154NYJ3 A0A1L8DYS7 A0A0L7RHQ2 A0A310SC29 A0A2A3EUE7 A0A088AGY3 A0A182Q368 D6WDP5 E2AFH3 A0A0C9RFJ1 A0A026WC95 A0A151XAS8 A0A195DHT8 A0A158P1Q5 A0A195EQR8 A0A195BSR1 A0A195CMA1 A0A1Y1MCI8 A0A067R932 E0VL53 A0A1B6HBE2 A0A1B6MNW0 A0A2M4D0Q4 K7IX55 A0A1B6DK62 W8CCJ2 A0A2M3Z8M5 A0A0K8VIY2 N6UM55 A0A0A1XMJ5 A0A034WI04 A0A0P4VQG4 A0A023FBP6 A0A0M4E862 R4WDZ4 B4H402 A0A0A9WQ72 B4KWK9 B3NF21 Q29E36 B4IYW4 B4LEC0 B4PCW2 B3M7F2 A0A0L0CI52 A0A1W4W1V7 A0A3B0J5A4 B4QP12 Q9VZW0 B4N6I1 A0A224XK94 A0A0M9A1I0 A0A1I8N340 A0A1A9XHI0 A0A1B0BHV1 B4HTJ0 A0A336LKV8 A0A1A9VED7 A0A1A9Z4L0 A0A2H8TZI5 J9K841 A0A1B0FA55 A0A1A9W7I7 A0A1B0AIY1 A0A1B0B759 A0A1B0G1S5 A0A1A9Y1D9 A0A0M3QZI6 A0A0L0C859 A0A2S2R5T4 A0A0A1WZY3 A0A0A1WFF0 A0A0A1WJ54 B3NUD3 Q9VYY8
Q16YK7 U5ESN1 A0A1L8DZB5 A0A2C9GPF5 A0A182UZW9 Q7QIP4 A0A182ILN7 A0A182P132 A0A2J7QKZ7 A0A182JTG2 A0A1B0CMB3 A0A2M4BHW4 A0A2M3Z7Q2 W5JA25 A0A154NYJ3 A0A1L8DYS7 A0A0L7RHQ2 A0A310SC29 A0A2A3EUE7 A0A088AGY3 A0A182Q368 D6WDP5 E2AFH3 A0A0C9RFJ1 A0A026WC95 A0A151XAS8 A0A195DHT8 A0A158P1Q5 A0A195EQR8 A0A195BSR1 A0A195CMA1 A0A1Y1MCI8 A0A067R932 E0VL53 A0A1B6HBE2 A0A1B6MNW0 A0A2M4D0Q4 K7IX55 A0A1B6DK62 W8CCJ2 A0A2M3Z8M5 A0A0K8VIY2 N6UM55 A0A0A1XMJ5 A0A034WI04 A0A0P4VQG4 A0A023FBP6 A0A0M4E862 R4WDZ4 B4H402 A0A0A9WQ72 B4KWK9 B3NF21 Q29E36 B4IYW4 B4LEC0 B4PCW2 B3M7F2 A0A0L0CI52 A0A1W4W1V7 A0A3B0J5A4 B4QP12 Q9VZW0 B4N6I1 A0A224XK94 A0A0M9A1I0 A0A1I8N340 A0A1A9XHI0 A0A1B0BHV1 B4HTJ0 A0A336LKV8 A0A1A9VED7 A0A1A9Z4L0 A0A2H8TZI5 J9K841 A0A1B0FA55 A0A1A9W7I7 A0A1B0AIY1 A0A1B0B759 A0A1B0G1S5 A0A1A9Y1D9 A0A0M3QZI6 A0A0L0C859 A0A2S2R5T4 A0A0A1WZY3 A0A0A1WFF0 A0A0A1WJ54 B3NUD3 Q9VYY8
PDB
3VZB
E-value=6.38269e-38,
Score=397
Ontologies
PATHWAY
GO
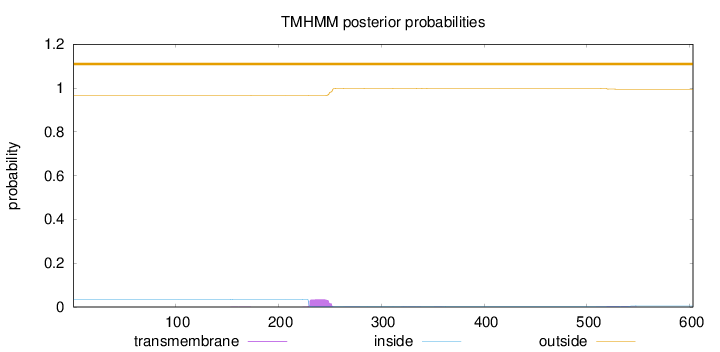
Topology
Length:
603
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.827150000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03510
outside
1 - 603
Population Genetic Test Statistics
Pi
229.534736
Theta
192.265781
Tajima's D
0.656998
CLR
0.033539
CSRT
0.564721763911804
Interpretation
Possibly Positive selection
