Gene
KWMTBOMO06479
Pre Gene Modal
BGIBMGA011859
Annotation
PREDICTED:_kelch-like_protein_10_[Amyelois_transitella]
Full name
Kelch-like protein 10
Location in the cell
PlasmaMembrane Reliability : 2.815
Sequence
CDS
ATGGAATTTAATACGAAACATAAAAAATTAACAAAAGCCAAAAATTCTGTACGTACGCCTTCAATTGCGAGATCACGTTTAAACAAAAAGAAAATTATCACGAGACAAAGAAAATGTGCGTGTCTTCCTGAAAATTATTCTGTGGTAGAATTCCCAGCAATTTGGAATGAACTACGACAAAACGGTCAGCTCTGTGATGGAACGATAATGTGTAGGGATTTGAAAGCAATAGGTGTTCATCGCGCCATACTATCAGCAGTTAGCCCATATTTTAAAGCTATATTTATAAACTCTTTAAACAAAGGTGAACCTGAGGAAACTAAAATATTTGTAGATGTTCCTAGTTTCTATATGAATCTAATACTGGATTACGCTTATACTGGAACTTGTAAGGTTACAGCAGAGAATGTTGAGTATCTTCTGCCATACGCAGACAAATTCGATGTTGTGGGAGTTATCCAACTTTGTTGCCAGTACTTACTGCAAGAATTAAAACCTCATAATTGCCTAGGAATATTTAAATTTGCTCGATATTATTTTTGCGGTGAATTAGAAAAGAAGGGGAAAATGTTTATAAGACAGCACTTTAATAAGATACTAAAGGAATGTAATGAATTTAAATCGCTTACTTTCGAAGAATTGGAAGACATTTTACGAGATGACGAACTGAACGTTAAAAATGAAGAAATTGTTTTTCAAGCTTTAAAGACTTGGGTCGAACATGACTTAGAAAACAGGAGAAAGTACATTCCGTCATTGCTTTCATGCGTTAGATTTGGTCACATAAGTTTTAAATATTTCAAATCAAAAATATTACAATGGCAGCCAGTGGTTGATGATGTGAAATGCCAAGAAGCGTTATACCCAGCAGTGGTGTTTTTAACATTACTAGATTCAAGACCTGGTATGGAAGCAGATTTAAACGATCCACTGGCGCGACCTAGAATTCCTTTTGAAATACTTTTTGCTGTTGGAGGATGGAGTGCTGGTAGTCCTACGAGCTTTGTTGAAACATACGATACAAGAGCTGACCGCTGGTTTCTTTCAATTCACATGGACCTTACTCCACGTGCTTATCACGGCCTCTGCACACTGAACAATCTGATTTATATGATCGGAGGTTTTGATGGTAGTGACCATTTTAATACGGTACGTTGTTACGACCCTGTCACTAATACGTGGCACGAACGCGCCTGTATGTATCAAGCCAGATGCTACGTCAGCGTTGTCGTGCACGACGGTATGATATACGCACTTGGAGGATATAATGGACGTACTCGTATGGCTTCAGTAGAACGGTACTATCCTGACAGAAATCAATGGGAGATGACCACTCCAATGAATAAACAGCGCTCAGACGCTAGCGCGGCATCACTAGGTGGCAAGATTTATATAGTGGGCGGTTTTAATGGACAAGAAGTATTGAGTTCTGCAGAAGTCTTTGACCCAGAGACGCGTCAATGGAGTTTCATCAGGTCGATGCTCAGTCCTCGCTCGGGAGTTAGTCTCATCGCCTACAGGGAATGCTTATTCGCGCTAGGTGGCTTCAATGGATACAGTCGTCTTAATACTGGTGAGAAATTTAATCCACAACGCGGCGGCGAATGGCAGGAAGTAACGGAAATGTTTAGTGCAAGAAGTAATTTCGCCACGGTTCTGTTGGACGATATGATTTTTGTCATCGGAGGATTTAATGGGTCCACAACAATTCCTCTCGTGGAATGCTACGATGGAGATACTCTAGAGTGGTACGACGCCGCTCCAATGAACTTGAATCGTTCAGCGCTCAGTGCTTGTGTTCTTGCCGGTTTACCGAACGCGCGTTCCTTCTCTTATTTGGCGAAAGCTGTGCCTGCCGCTGGTGCTGATCAAGCTCCTCAATCCTAA
Protein
MEFNTKHKKLTKAKNSVRTPSIARSRLNKKKIITRQRKCACLPENYSVVEFPAIWNELRQNGQLCDGTIMCRDLKAIGVHRAILSAVSPYFKAIFINSLNKGEPEETKIFVDVPSFYMNLILDYAYTGTCKVTAENVEYLLPYADKFDVVGVIQLCCQYLLQELKPHNCLGIFKFARYYFCGELEKKGKMFIRQHFNKILKECNEFKSLTFEELEDILRDDELNVKNEEIVFQALKTWVEHDLENRRKYIPSLLSCVRFGHISFKYFKSKILQWQPVVDDVKCQEALYPAVVFLTLLDSRPGMEADLNDPLARPRIPFEILFAVGGWSAGSPTSFVETYDTRADRWFLSIHMDLTPRAYHGLCTLNNLIYMIGGFDGSDHFNTVRCYDPVTNTWHERACMYQARCYVSVVVHDGMIYALGGYNGRTRMASVERYYPDRNQWEMTTPMNKQRSDASAASLGGKIYIVGGFNGQEVLSSAEVFDPETRQWSFIRSMLSPRSGVSLIAYRECLFALGGFNGYSRLNTGEKFNPQRGGEWQEVTEMFSARSNFATVLLDDMIFVIGGFNGSTTIPLVECYDGDTLEWYDAAPMNLNRSALSACVLAGLPNARSFSYLAKAVPAAGADQAPQS
Summary
Description
May be a substrate-specific adapter of a CUL3-based E3 ubiquitin-protein ligase complex which mediates the ubiquitination and subsequent proteasomal degradation of target proteins during spermatogenesis. Required for male fertility.
Subunit
Self-associates. Interacts with CUL3; indicative for the participation in an E3 ubiquitin ligase complex (By similarity).
Self-associates (By similarity). Interacts with CUL3; indicative for the participation in an E3 ubiquitin ligase complex.
Self-associates (Probable). Interacts with CUL3; indicative for the participation in an E3 ubiquitin ligase complex (By similarity).
Self-associates (By similarity). Interacts with CUL3; indicative for the participation in an E3 ubiquitin ligase complex.
Self-associates (Probable). Interacts with CUL3; indicative for the participation in an E3 ubiquitin ligase complex (By similarity).
Miscellaneous
Does not seem to associate with actin.
Keywords
Complete proteome
Cytoplasm
Differentiation
Kelch repeat
Phosphoprotein
Reference proteome
Repeat
Spermatogenesis
Ubl conjugation pathway
Disease mutation
Feature
chain Kelch-like protein 10
sequence variant In SPGF11; results in impaired self-association; dbSNP:rs116420871.
sequence variant In SPGF11; results in impaired self-association; dbSNP:rs116420871.
Uniprot
H9JQQ3
A0A2W1BU36
A0A2H1WL14
A0A2A4JPE0
A0A3S2P6P5
A0A194PWT1
+ More
A0A212FP59 A0A0L7KNF5 A0A195CUZ8 A0A151X7K8 A0A195DLS5 A0A088A511 A0A195BKG8 A0A158NAS0 A0A195FR76 A0A154PC83 A0A0L7QTB4 A0A026WMW5 A0A2P8ZKG0 A0A3L8D7J3 A0A1W4X0Q7 E0V9G0 A0A1W4WPP8 A0A0C9RC58 E9IXQ8 A0A139WKL2 A0A139WKC4 A0A1Y1KKP3 T1IMJ6 N6TZP3 A0A0M9A696 A0A226EDW8 A0A2J7R646 A0A2C9KFA9 A0A2C9KF84 A0A3S1AGY3 A0A210QDN8 V4B2M3 A0A210QKN9 V4BZ38 K1QZM1 A0A210QRR8 E0VNX1 A0A0N1PI19 K1R9Z0 A0A1W4X0E7 T1HSE5 A0A151PC46 A0A2A3EI13 A0A088A9H5 H2ZWD1 A0A224X953 A0A195D368 K7FXG8 E2AY31 A0A1B6IL54 A0A0L8HH00 A0A2C9JDU9 A0A151WTN2 A0A195DM84 A0A383ZRK8 A0A0L7RH03 A0A195FJR8 A0A195BTD2 A0A158P2D9 A0A2K5YXL9 A0A1S3S5E1 D2HIS0 H2NU21 F1Q2G6 A0A3Q7RC13 A0A0A9YPI4 A0A2R9AIC9 H2QD07 A0A1Y1NHD7 L8IEH6 F1MBZ1 G3SZ77 G1PJA9 A0A1S3GWH7 I3M727 B2RTD7 A0A2Y9EBS7 Q6JEL3 Q9D5V2 A0A1S3AIB7 H0WGF8 A0A1U7QWK6 G3R3Q6 A0A140VJM8 Q6JEL2 A0A2K5Q0A6 A0A2K6S0A5 A0A384CXH6 A0A287BE53 F6XWF4 A0A3Q7NGP2 A0A2Y9JWC6 A0A3P4MXN4
A0A212FP59 A0A0L7KNF5 A0A195CUZ8 A0A151X7K8 A0A195DLS5 A0A088A511 A0A195BKG8 A0A158NAS0 A0A195FR76 A0A154PC83 A0A0L7QTB4 A0A026WMW5 A0A2P8ZKG0 A0A3L8D7J3 A0A1W4X0Q7 E0V9G0 A0A1W4WPP8 A0A0C9RC58 E9IXQ8 A0A139WKL2 A0A139WKC4 A0A1Y1KKP3 T1IMJ6 N6TZP3 A0A0M9A696 A0A226EDW8 A0A2J7R646 A0A2C9KFA9 A0A2C9KF84 A0A3S1AGY3 A0A210QDN8 V4B2M3 A0A210QKN9 V4BZ38 K1QZM1 A0A210QRR8 E0VNX1 A0A0N1PI19 K1R9Z0 A0A1W4X0E7 T1HSE5 A0A151PC46 A0A2A3EI13 A0A088A9H5 H2ZWD1 A0A224X953 A0A195D368 K7FXG8 E2AY31 A0A1B6IL54 A0A0L8HH00 A0A2C9JDU9 A0A151WTN2 A0A195DM84 A0A383ZRK8 A0A0L7RH03 A0A195FJR8 A0A195BTD2 A0A158P2D9 A0A2K5YXL9 A0A1S3S5E1 D2HIS0 H2NU21 F1Q2G6 A0A3Q7RC13 A0A0A9YPI4 A0A2R9AIC9 H2QD07 A0A1Y1NHD7 L8IEH6 F1MBZ1 G3SZ77 G1PJA9 A0A1S3GWH7 I3M727 B2RTD7 A0A2Y9EBS7 Q6JEL3 Q9D5V2 A0A1S3AIB7 H0WGF8 A0A1U7QWK6 G3R3Q6 A0A140VJM8 Q6JEL2 A0A2K5Q0A6 A0A2K6S0A5 A0A384CXH6 A0A287BE53 F6XWF4 A0A3Q7NGP2 A0A2Y9JWC6 A0A3P4MXN4
Pubmed
19121390
28756777
26354079
22118469
26227816
21347285
+ More
24508170 29403074 30249741 20566863 21282665 18362917 19820115 28004739 23537049 15562597 28812685 23254933 22992520 22293439 9215903 17381049 20798317 20010809 16341006 25401762 22722832 16136131 22751099 19393038 21993624 12040188 15489334 21183079 15136734 22673903 16141072 16162871 22398555 11181995 14702039 17047026 24813606 19892987
24508170 29403074 30249741 20566863 21282665 18362917 19820115 28004739 23537049 15562597 28812685 23254933 22992520 22293439 9215903 17381049 20798317 20010809 16341006 25401762 22722832 16136131 22751099 19393038 21993624 12040188 15489334 21183079 15136734 22673903 16141072 16162871 22398555 11181995 14702039 17047026 24813606 19892987
EMBL
BABH01012284
KZ149967
PZC76156.1
ODYU01009339
SOQ53697.1
NWSH01000980
+ More
PCG73282.1 RSAL01000010 RVE53719.1 KQ459590 KPI97219.1 AGBW02003646 OWR55515.1 JTDY01008346 KOB64611.1 KQ977276 KYN04498.1 KQ982446 KYQ56314.1 KQ980734 KYN13828.1 KQ976455 KYM85143.1 ADTU01010045 KQ981305 KYN42968.1 KQ434870 KZC09505.1 KQ414755 KOC61721.1 KK107153 EZA56991.1 PYGN01000029 PSN56988.1 QOIP01000012 RLU16269.1 DS234992 EEB10016.1 GBYB01014169 JAG83936.1 GL766762 EFZ14665.1 KQ971329 KYB28355.1 KYB28354.1 GEZM01080967 JAV61963.1 JH431067 APGK01045172 APGK01045173 KB741031 KI208367 KI208683 ENN74765.1 ERL95846.1 ERL95906.1 KQ435727 KOX78097.1 LNIX01000004 OXA55334.1 NEVH01006981 PNF36303.1 RQTK01000005 RUS91794.1 NEDP02004067 OWF46852.1 KB200701 ESP00682.1 NEDP02003184 OWF49315.1 KB201813 ESO94394.1 JH818378 EKC42522.1 NEDP02002269 OWF51388.1 DS235354 EEB15077.1 KQ461161 KPJ08012.1 EKC42523.1 ACPB03004813 AKHW03000499 KYO46686.1 KZ288254 PBC30839.1 AFYH01149238 AFYH01149239 GFTR01007560 JAW08866.1 KQ976885 KYN07355.1 AGCU01138943 GL443762 EFN61627.1 GECU01020049 JAS87657.1 KQ418269 KOF88050.1 KQ982753 KYQ51196.1 KYN13946.1 KQ414594 KOC70140.1 KQ981523 KYN40249.1 KQ976408 KYM91196.1 ADTU01007164 ADTU01007165 ACTA01012009 GL192892 EFB20106.1 ABGA01331353 ABGA01331354 ABGA01331355 NDHI03003706 PNJ07227.1 AAEX03006455 GBHO01010073 GBRD01001242 JAG33531.1 JAG64579.1 AJFE02044423 AJFE02044424 AJFE02044425 AJFE02044426 AJFE02044427 AJFE02044428 AACZ04038960 NBAG03000371 PNI33986.1 GEZM01002346 JAV97312.1 JH881506 ELR53939.1 AAPE02024581 AGTP01023477 BC139267 BC139268 CH466662 AAI39268.1 EDL02558.1 AY495338 BC085842 AY495337 AK006288 AK014906 AK077202 AAQR03042772 AAQR03042773 CABD030037877 HM005417 CH471152 AEE61015.1 EAW60774.1 AY495339 AK057224 AEMK02000080 CYRY02015514 VCW85495.1
PCG73282.1 RSAL01000010 RVE53719.1 KQ459590 KPI97219.1 AGBW02003646 OWR55515.1 JTDY01008346 KOB64611.1 KQ977276 KYN04498.1 KQ982446 KYQ56314.1 KQ980734 KYN13828.1 KQ976455 KYM85143.1 ADTU01010045 KQ981305 KYN42968.1 KQ434870 KZC09505.1 KQ414755 KOC61721.1 KK107153 EZA56991.1 PYGN01000029 PSN56988.1 QOIP01000012 RLU16269.1 DS234992 EEB10016.1 GBYB01014169 JAG83936.1 GL766762 EFZ14665.1 KQ971329 KYB28355.1 KYB28354.1 GEZM01080967 JAV61963.1 JH431067 APGK01045172 APGK01045173 KB741031 KI208367 KI208683 ENN74765.1 ERL95846.1 ERL95906.1 KQ435727 KOX78097.1 LNIX01000004 OXA55334.1 NEVH01006981 PNF36303.1 RQTK01000005 RUS91794.1 NEDP02004067 OWF46852.1 KB200701 ESP00682.1 NEDP02003184 OWF49315.1 KB201813 ESO94394.1 JH818378 EKC42522.1 NEDP02002269 OWF51388.1 DS235354 EEB15077.1 KQ461161 KPJ08012.1 EKC42523.1 ACPB03004813 AKHW03000499 KYO46686.1 KZ288254 PBC30839.1 AFYH01149238 AFYH01149239 GFTR01007560 JAW08866.1 KQ976885 KYN07355.1 AGCU01138943 GL443762 EFN61627.1 GECU01020049 JAS87657.1 KQ418269 KOF88050.1 KQ982753 KYQ51196.1 KYN13946.1 KQ414594 KOC70140.1 KQ981523 KYN40249.1 KQ976408 KYM91196.1 ADTU01007164 ADTU01007165 ACTA01012009 GL192892 EFB20106.1 ABGA01331353 ABGA01331354 ABGA01331355 NDHI03003706 PNJ07227.1 AAEX03006455 GBHO01010073 GBRD01001242 JAG33531.1 JAG64579.1 AJFE02044423 AJFE02044424 AJFE02044425 AJFE02044426 AJFE02044427 AJFE02044428 AACZ04038960 NBAG03000371 PNI33986.1 GEZM01002346 JAV97312.1 JH881506 ELR53939.1 AAPE02024581 AGTP01023477 BC139267 BC139268 CH466662 AAI39268.1 EDL02558.1 AY495338 BC085842 AY495337 AK006288 AK014906 AK077202 AAQR03042772 AAQR03042773 CABD030037877 HM005417 CH471152 AEE61015.1 EAW60774.1 AY495339 AK057224 AEMK02000080 CYRY02015514 VCW85495.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000037510
+ More
UP000078542 UP000075809 UP000078492 UP000005203 UP000078540 UP000005205 UP000078541 UP000076502 UP000053825 UP000053097 UP000245037 UP000279307 UP000192223 UP000009046 UP000007266 UP000019118 UP000030742 UP000053105 UP000198287 UP000235965 UP000076420 UP000271974 UP000242188 UP000030746 UP000005408 UP000053240 UP000015103 UP000050525 UP000242457 UP000008672 UP000007267 UP000000311 UP000053454 UP000261681 UP000233140 UP000087266 UP000008912 UP000001595 UP000002254 UP000286640 UP000240080 UP000002277 UP000009136 UP000007646 UP000001074 UP000081671 UP000005215 UP000248480 UP000002494 UP000000589 UP000079721 UP000005225 UP000189706 UP000001519 UP000005640 UP000233040 UP000233220 UP000261680 UP000291021 UP000008227 UP000002281 UP000286641 UP000248482
UP000078542 UP000075809 UP000078492 UP000005203 UP000078540 UP000005205 UP000078541 UP000076502 UP000053825 UP000053097 UP000245037 UP000279307 UP000192223 UP000009046 UP000007266 UP000019118 UP000030742 UP000053105 UP000198287 UP000235965 UP000076420 UP000271974 UP000242188 UP000030746 UP000005408 UP000053240 UP000015103 UP000050525 UP000242457 UP000008672 UP000007267 UP000000311 UP000053454 UP000261681 UP000233140 UP000087266 UP000008912 UP000001595 UP000002254 UP000286640 UP000240080 UP000002277 UP000009136 UP000007646 UP000001074 UP000081671 UP000005215 UP000248480 UP000002494 UP000000589 UP000079721 UP000005225 UP000189706 UP000001519 UP000005640 UP000233040 UP000233220 UP000261680 UP000291021 UP000008227 UP000002281 UP000286641 UP000248482
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JQQ3
A0A2W1BU36
A0A2H1WL14
A0A2A4JPE0
A0A3S2P6P5
A0A194PWT1
+ More
A0A212FP59 A0A0L7KNF5 A0A195CUZ8 A0A151X7K8 A0A195DLS5 A0A088A511 A0A195BKG8 A0A158NAS0 A0A195FR76 A0A154PC83 A0A0L7QTB4 A0A026WMW5 A0A2P8ZKG0 A0A3L8D7J3 A0A1W4X0Q7 E0V9G0 A0A1W4WPP8 A0A0C9RC58 E9IXQ8 A0A139WKL2 A0A139WKC4 A0A1Y1KKP3 T1IMJ6 N6TZP3 A0A0M9A696 A0A226EDW8 A0A2J7R646 A0A2C9KFA9 A0A2C9KF84 A0A3S1AGY3 A0A210QDN8 V4B2M3 A0A210QKN9 V4BZ38 K1QZM1 A0A210QRR8 E0VNX1 A0A0N1PI19 K1R9Z0 A0A1W4X0E7 T1HSE5 A0A151PC46 A0A2A3EI13 A0A088A9H5 H2ZWD1 A0A224X953 A0A195D368 K7FXG8 E2AY31 A0A1B6IL54 A0A0L8HH00 A0A2C9JDU9 A0A151WTN2 A0A195DM84 A0A383ZRK8 A0A0L7RH03 A0A195FJR8 A0A195BTD2 A0A158P2D9 A0A2K5YXL9 A0A1S3S5E1 D2HIS0 H2NU21 F1Q2G6 A0A3Q7RC13 A0A0A9YPI4 A0A2R9AIC9 H2QD07 A0A1Y1NHD7 L8IEH6 F1MBZ1 G3SZ77 G1PJA9 A0A1S3GWH7 I3M727 B2RTD7 A0A2Y9EBS7 Q6JEL3 Q9D5V2 A0A1S3AIB7 H0WGF8 A0A1U7QWK6 G3R3Q6 A0A140VJM8 Q6JEL2 A0A2K5Q0A6 A0A2K6S0A5 A0A384CXH6 A0A287BE53 F6XWF4 A0A3Q7NGP2 A0A2Y9JWC6 A0A3P4MXN4
A0A212FP59 A0A0L7KNF5 A0A195CUZ8 A0A151X7K8 A0A195DLS5 A0A088A511 A0A195BKG8 A0A158NAS0 A0A195FR76 A0A154PC83 A0A0L7QTB4 A0A026WMW5 A0A2P8ZKG0 A0A3L8D7J3 A0A1W4X0Q7 E0V9G0 A0A1W4WPP8 A0A0C9RC58 E9IXQ8 A0A139WKL2 A0A139WKC4 A0A1Y1KKP3 T1IMJ6 N6TZP3 A0A0M9A696 A0A226EDW8 A0A2J7R646 A0A2C9KFA9 A0A2C9KF84 A0A3S1AGY3 A0A210QDN8 V4B2M3 A0A210QKN9 V4BZ38 K1QZM1 A0A210QRR8 E0VNX1 A0A0N1PI19 K1R9Z0 A0A1W4X0E7 T1HSE5 A0A151PC46 A0A2A3EI13 A0A088A9H5 H2ZWD1 A0A224X953 A0A195D368 K7FXG8 E2AY31 A0A1B6IL54 A0A0L8HH00 A0A2C9JDU9 A0A151WTN2 A0A195DM84 A0A383ZRK8 A0A0L7RH03 A0A195FJR8 A0A195BTD2 A0A158P2D9 A0A2K5YXL9 A0A1S3S5E1 D2HIS0 H2NU21 F1Q2G6 A0A3Q7RC13 A0A0A9YPI4 A0A2R9AIC9 H2QD07 A0A1Y1NHD7 L8IEH6 F1MBZ1 G3SZ77 G1PJA9 A0A1S3GWH7 I3M727 B2RTD7 A0A2Y9EBS7 Q6JEL3 Q9D5V2 A0A1S3AIB7 H0WGF8 A0A1U7QWK6 G3R3Q6 A0A140VJM8 Q6JEL2 A0A2K5Q0A6 A0A2K6S0A5 A0A384CXH6 A0A287BE53 F6XWF4 A0A3Q7NGP2 A0A2Y9JWC6 A0A3P4MXN4
PDB
6GY5
E-value=1.80452e-49,
Score=497
Ontologies
GO
Topology
Subcellular location
Cytoplasm
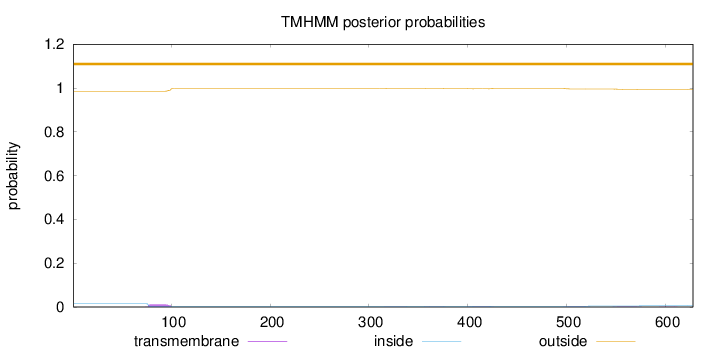
Length:
628
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.41963
Exp number, first 60 AAs:
0.00074
Total prob of N-in:
0.01492
outside
1 - 628
Population Genetic Test Statistics
Pi
233.443947
Theta
186.532085
Tajima's D
0.534471
CLR
0.531301
CSRT
0.528223588820559
Interpretation
Uncertain
