Pre Gene Modal
BGIBMGA011860
Annotation
PREDICTED:_15-hydroxyprostaglandin_dehydrogenase_[NAD(+)]-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.84
Sequence
CDS
ATGGACTTAAAAGGAAAGGTTGCATTGATAACGGGCGCGGCTTCCGGCATCGGCTTAGCCTATAGCGAAGAATTATTAAAACAAGGTGCTAAGGTTTCCCTATGCGACATCGATTCGGAGCGCGGGGAACAAGTAACAGAAAAGCTCGCTTCAAAATATGGCCGAAAAAACGTTTTGTTTTGTCGATTAGATGTTACTGACTATCCACAATTTGAAGAGGCATTTGTGATGACAGTAGAGGAGTTCAACCGGATTGACATTGTCATTAATAATGCTGGCTTCATGAACGACAGGCTTTGGGAACTCGAAGTCGATGTTAATTTGAATGGCGTCATCAGAGGTACGCTGCTCGCATATCGGTTTATGGGGAAGGATCGCGGAGGGCAAGGCGGTGTCATCGTGAACACGGCCTCTACAGCATTTGCGAGACCTCAAGTGTCGACTCCGATCTACACAGCAACGAACTACGCTATTGTTGGCCTCACCAGAGCTTACGGGGATCAGTACCACGTCAATATGACCGGAGTGCGATGCCTCGTCGTGTGCCCTGGACTCACGGACAGTGGTTTGGTGAAGGATCTTGACAAGCAGCTTATGTCTTCAAAATACGAGACTGCTTGGCTAGGGGACATTACAGATTCTAAAATGCAGAGCTCTGATCACGTTGGGAGAGCCCTGGTGGATGTACTCGCGAAAGGTCAAAGCGGCTCAGTTTGGATTGTCGAAAACGCTCAGCCGGCTAGGGAGTGGCGCGGCTAA
Protein
MDLKGKVALITGAASGIGLAYSEELLKQGAKVSLCDIDSERGEQVTEKLASKYGRKNVLFCRLDVTDYPQFEEAFVMTVEEFNRIDIVINNAGFMNDRLWELEVDVNLNGVIRGTLLAYRFMGKDRGGQGGVIVNTASTAFARPQVSTPIYTATNYAIVGLTRAYGDQYHVNMTGVRCLVVCPGLTDSGLVKDLDKQLMSSKYETAWLGDITDSKMQSSDHVGRALVDVLAKGQSGSVWIVENAQPAREWRG
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
H9JQQ4
A0A2A4JPA8
A0A3S2NQL5
A0A2H1WL48
A0A2W1BRA8
A0A0N1INL6
+ More
A0A194Q1E5 A0A212FP41 A0A067QWG1 A0A2J7QGZ8 K7IM82 A0A0J7L1B2 E2A713 A0A232ERU5 A0A195B3G3 E9I9W8 A0A195E5P2 F4W475 A0A151X250 A0A195CJP6 A0A195EUB2 A0A158NP60 A0A0C9RAC9 E2B6N1 A0A026WSD7 A0A154PC81 A0A088A4Z7 A0A1B6DW76 D6WGE7 A0A310S8E6 A0A1W4WPT7 A0A2H8TFE7 J9JKN0 A0A2S2NV02 E0VAP2 J9HSC3 A0A1B0CRR2 A0A1W7R8C8 A0A1B0D1S5 J3JUA4 A0A069DQD9 A0A224XI03 A0A182QF31 A0A182VRS1 A0A182LSI1 A0A182NLI5 A0A146M7A4 A0A0A9Z8Q7 A0A0M9A8L6 A0A182XV05 A0A182RPC3 A0A182WSW9 A0A182V3V9 A0A182LA81 A0A182TSI2 A0A182HLS3 A0A2S2R4Y7 Q7PY55 A0A182SMS8 A0A1Y1KU79 A0A2A3ECA0 W5JFH6 A0A182NZC9 A0A1B0GNT7 A0A182FNL1 A0A1W4X0U4 A0A0L7QSV8 T1HQ12 A0A182KA05 A0A182JE34 A0A067R8Z3 A0A0T6B6V3 A0A1W4X358 A0A1Y1MAW1 A0A2P8Z2R9 A0A1W4XDZ2 A0A2J7QR46 A0A2P8Y4B5 D6WT10 A0A140KPR8 D6WG91 A0A2P8ZLP3 R4FLI9 Q6RUR3 A0A2J7QR36 A0A1W4X3A0 A0A084W563 A0A1Y1N5U0 A0A151WLU8 A0A151XEI7 A0A023F7A3 A0A067R8A9 A0A1Y1N8U2 V5GSR5 A0A224XI56 A0A1Y1MYW2 A0A1Y1N8K6 A0A154PE73 A0A023FB52 A0A224XI78
A0A194Q1E5 A0A212FP41 A0A067QWG1 A0A2J7QGZ8 K7IM82 A0A0J7L1B2 E2A713 A0A232ERU5 A0A195B3G3 E9I9W8 A0A195E5P2 F4W475 A0A151X250 A0A195CJP6 A0A195EUB2 A0A158NP60 A0A0C9RAC9 E2B6N1 A0A026WSD7 A0A154PC81 A0A088A4Z7 A0A1B6DW76 D6WGE7 A0A310S8E6 A0A1W4WPT7 A0A2H8TFE7 J9JKN0 A0A2S2NV02 E0VAP2 J9HSC3 A0A1B0CRR2 A0A1W7R8C8 A0A1B0D1S5 J3JUA4 A0A069DQD9 A0A224XI03 A0A182QF31 A0A182VRS1 A0A182LSI1 A0A182NLI5 A0A146M7A4 A0A0A9Z8Q7 A0A0M9A8L6 A0A182XV05 A0A182RPC3 A0A182WSW9 A0A182V3V9 A0A182LA81 A0A182TSI2 A0A182HLS3 A0A2S2R4Y7 Q7PY55 A0A182SMS8 A0A1Y1KU79 A0A2A3ECA0 W5JFH6 A0A182NZC9 A0A1B0GNT7 A0A182FNL1 A0A1W4X0U4 A0A0L7QSV8 T1HQ12 A0A182KA05 A0A182JE34 A0A067R8Z3 A0A0T6B6V3 A0A1W4X358 A0A1Y1MAW1 A0A2P8Z2R9 A0A1W4XDZ2 A0A2J7QR46 A0A2P8Y4B5 D6WT10 A0A140KPR8 D6WG91 A0A2P8ZLP3 R4FLI9 Q6RUR3 A0A2J7QR36 A0A1W4X3A0 A0A084W563 A0A1Y1N5U0 A0A151WLU8 A0A151XEI7 A0A023F7A3 A0A067R8A9 A0A1Y1N8U2 V5GSR5 A0A224XI56 A0A1Y1MYW2 A0A1Y1N8K6 A0A154PE73 A0A023FB52 A0A224XI78
Pubmed
19121390
28756777
26354079
22118469
24845553
20075255
+ More
20798317 28648823 21282665 21719571 21347285 24508170 30249741 18362917 19820115 20566863 17510324 22516182 23537049 26334808 26823975 25401762 25244985 20966253 12364791 14747013 17210077 28004739 20920257 23761445 29403074 24438588 25474469
20798317 28648823 21282665 21719571 21347285 24508170 30249741 18362917 19820115 20566863 17510324 22516182 23537049 26334808 26823975 25401762 25244985 20966253 12364791 14747013 17210077 28004739 20920257 23761445 29403074 24438588 25474469
EMBL
BABH01012284
BABH01012285
NWSH01000980
PCG73283.1
RSAL01000010
RVE53718.1
+ More
ODYU01009339 SOQ53696.1 KZ149967 PZC76155.1 KQ461161 KPJ08013.1 KQ459590 KPI97220.1 AGBW02003646 OWR55516.1 KK852879 KDR14474.1 NEVH01014359 PNF27833.1 LBMM01001364 KMQ96441.1 GL437252 EFN70745.1 NNAY01002548 OXU21079.1 KQ976625 KYM79033.1 GL761846 EFZ22664.1 KQ979609 KYN20169.1 GL887491 EGI71071.1 KQ982585 KYQ54300.1 KQ977642 KYN00938.1 KQ981979 KYN31469.1 ADTU01001531 GBYB01013389 JAG83156.1 GL446000 EFN88615.1 KK107119 QOIP01000009 EZA58571.1 RLU18590.1 KQ434870 KZC09506.1 GEDC01007377 JAS29921.1 KQ971329 EFA01116.1 KQ771134 OAD52541.1 GFXV01001021 MBW12826.1 ABLF02032246 ABLF02032258 GGMR01008279 MBY20898.1 DS235012 EEB10448.1 CH477284 EJY57503.1 AJWK01025142 GEHC01000259 JAV47386.1 AJVK01022288 AJVK01022289 AJVK01022290 APGK01049722 BT126817 KB741156 KB631941 AEE61779.1 ENN73601.1 ERL87393.1 GBGD01002606 JAC86283.1 GFTR01004300 JAW12126.1 AXCN02000138 AXCM01000496 GDHC01004189 JAQ14440.1 GBHO01001997 JAG41607.1 KQ435727 KOX78099.1 APCN01000911 GGMS01015878 MBY85081.1 AAAB01008987 EAA00929.4 GEZM01076510 JAV63760.1 KZ288303 PBC28806.1 ADMH02001291 ETN63127.1 AJVK01029793 KQ414755 KOC61722.1 ACPB03013617 KK852855 KDR14911.1 LJIG01009455 KRT83046.1 GEZM01036109 GEZM01036108 JAV82969.1 PYGN01000223 PSN50783.1 NEVH01011985 PNF31053.1 PYGN01000947 PSN39080.1 KQ971354 EFA05875.1 LN794625 CEO43692.1 KQ971328 EFA00962.1 PYGN01000021 PSN57413.1 ACPB03004191 GAHY01001368 JAA76142.1 AY491503 AAR84629.1 PNF31051.1 ATLV01020510 KE525302 KFB45357.1 GEZM01011996 JAV93302.1 KQ982944 KYQ48859.1 KQ982254 KYQ58773.1 GBBI01001633 JAC17079.1 KK852815 KDR15798.1 GEZM01012000 GEZM01011999 GEZM01011998 GEZM01011997 GEZM01011995 GEZM01011994 GEZM01011993 JAV93300.1 GALX01003804 JAB64662.1 GFTR01004259 JAW12167.1 GEZM01023251 JAV88537.1 GEZM01009878 JAV94252.1 KZC09510.1 GBBI01000005 JAC18707.1 GFTR01004241 JAW12185.1
ODYU01009339 SOQ53696.1 KZ149967 PZC76155.1 KQ461161 KPJ08013.1 KQ459590 KPI97220.1 AGBW02003646 OWR55516.1 KK852879 KDR14474.1 NEVH01014359 PNF27833.1 LBMM01001364 KMQ96441.1 GL437252 EFN70745.1 NNAY01002548 OXU21079.1 KQ976625 KYM79033.1 GL761846 EFZ22664.1 KQ979609 KYN20169.1 GL887491 EGI71071.1 KQ982585 KYQ54300.1 KQ977642 KYN00938.1 KQ981979 KYN31469.1 ADTU01001531 GBYB01013389 JAG83156.1 GL446000 EFN88615.1 KK107119 QOIP01000009 EZA58571.1 RLU18590.1 KQ434870 KZC09506.1 GEDC01007377 JAS29921.1 KQ971329 EFA01116.1 KQ771134 OAD52541.1 GFXV01001021 MBW12826.1 ABLF02032246 ABLF02032258 GGMR01008279 MBY20898.1 DS235012 EEB10448.1 CH477284 EJY57503.1 AJWK01025142 GEHC01000259 JAV47386.1 AJVK01022288 AJVK01022289 AJVK01022290 APGK01049722 BT126817 KB741156 KB631941 AEE61779.1 ENN73601.1 ERL87393.1 GBGD01002606 JAC86283.1 GFTR01004300 JAW12126.1 AXCN02000138 AXCM01000496 GDHC01004189 JAQ14440.1 GBHO01001997 JAG41607.1 KQ435727 KOX78099.1 APCN01000911 GGMS01015878 MBY85081.1 AAAB01008987 EAA00929.4 GEZM01076510 JAV63760.1 KZ288303 PBC28806.1 ADMH02001291 ETN63127.1 AJVK01029793 KQ414755 KOC61722.1 ACPB03013617 KK852855 KDR14911.1 LJIG01009455 KRT83046.1 GEZM01036109 GEZM01036108 JAV82969.1 PYGN01000223 PSN50783.1 NEVH01011985 PNF31053.1 PYGN01000947 PSN39080.1 KQ971354 EFA05875.1 LN794625 CEO43692.1 KQ971328 EFA00962.1 PYGN01000021 PSN57413.1 ACPB03004191 GAHY01001368 JAA76142.1 AY491503 AAR84629.1 PNF31051.1 ATLV01020510 KE525302 KFB45357.1 GEZM01011996 JAV93302.1 KQ982944 KYQ48859.1 KQ982254 KYQ58773.1 GBBI01001633 JAC17079.1 KK852815 KDR15798.1 GEZM01012000 GEZM01011999 GEZM01011998 GEZM01011997 GEZM01011995 GEZM01011994 GEZM01011993 JAV93300.1 GALX01003804 JAB64662.1 GFTR01004259 JAW12167.1 GEZM01023251 JAV88537.1 GEZM01009878 JAV94252.1 KZC09510.1 GBBI01000005 JAC18707.1 GFTR01004241 JAW12185.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000027135 UP000235965 UP000002358 UP000036403 UP000000311 UP000215335 UP000078540 UP000078492 UP000007755 UP000075809 UP000078542 UP000078541 UP000005205 UP000008237 UP000053097 UP000279307 UP000076502 UP000005203 UP000007266 UP000192223 UP000007819 UP000009046 UP000008820 UP000092461 UP000092462 UP000019118 UP000030742 UP000075886 UP000075920 UP000075883 UP000075884 UP000053105 UP000076408 UP000075900 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000075901 UP000242457 UP000000673 UP000075885 UP000069272 UP000053825 UP000015103 UP000075881 UP000075880 UP000245037 UP000030765
UP000027135 UP000235965 UP000002358 UP000036403 UP000000311 UP000215335 UP000078540 UP000078492 UP000007755 UP000075809 UP000078542 UP000078541 UP000005205 UP000008237 UP000053097 UP000279307 UP000076502 UP000005203 UP000007266 UP000192223 UP000007819 UP000009046 UP000008820 UP000092461 UP000092462 UP000019118 UP000030742 UP000075886 UP000075920 UP000075883 UP000075884 UP000053105 UP000076408 UP000075900 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000007062 UP000075901 UP000242457 UP000000673 UP000075885 UP000069272 UP000053825 UP000015103 UP000075881 UP000075880 UP000245037 UP000030765
PRIDE
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JQQ4
A0A2A4JPA8
A0A3S2NQL5
A0A2H1WL48
A0A2W1BRA8
A0A0N1INL6
+ More
A0A194Q1E5 A0A212FP41 A0A067QWG1 A0A2J7QGZ8 K7IM82 A0A0J7L1B2 E2A713 A0A232ERU5 A0A195B3G3 E9I9W8 A0A195E5P2 F4W475 A0A151X250 A0A195CJP6 A0A195EUB2 A0A158NP60 A0A0C9RAC9 E2B6N1 A0A026WSD7 A0A154PC81 A0A088A4Z7 A0A1B6DW76 D6WGE7 A0A310S8E6 A0A1W4WPT7 A0A2H8TFE7 J9JKN0 A0A2S2NV02 E0VAP2 J9HSC3 A0A1B0CRR2 A0A1W7R8C8 A0A1B0D1S5 J3JUA4 A0A069DQD9 A0A224XI03 A0A182QF31 A0A182VRS1 A0A182LSI1 A0A182NLI5 A0A146M7A4 A0A0A9Z8Q7 A0A0M9A8L6 A0A182XV05 A0A182RPC3 A0A182WSW9 A0A182V3V9 A0A182LA81 A0A182TSI2 A0A182HLS3 A0A2S2R4Y7 Q7PY55 A0A182SMS8 A0A1Y1KU79 A0A2A3ECA0 W5JFH6 A0A182NZC9 A0A1B0GNT7 A0A182FNL1 A0A1W4X0U4 A0A0L7QSV8 T1HQ12 A0A182KA05 A0A182JE34 A0A067R8Z3 A0A0T6B6V3 A0A1W4X358 A0A1Y1MAW1 A0A2P8Z2R9 A0A1W4XDZ2 A0A2J7QR46 A0A2P8Y4B5 D6WT10 A0A140KPR8 D6WG91 A0A2P8ZLP3 R4FLI9 Q6RUR3 A0A2J7QR36 A0A1W4X3A0 A0A084W563 A0A1Y1N5U0 A0A151WLU8 A0A151XEI7 A0A023F7A3 A0A067R8A9 A0A1Y1N8U2 V5GSR5 A0A224XI56 A0A1Y1MYW2 A0A1Y1N8K6 A0A154PE73 A0A023FB52 A0A224XI78
A0A194Q1E5 A0A212FP41 A0A067QWG1 A0A2J7QGZ8 K7IM82 A0A0J7L1B2 E2A713 A0A232ERU5 A0A195B3G3 E9I9W8 A0A195E5P2 F4W475 A0A151X250 A0A195CJP6 A0A195EUB2 A0A158NP60 A0A0C9RAC9 E2B6N1 A0A026WSD7 A0A154PC81 A0A088A4Z7 A0A1B6DW76 D6WGE7 A0A310S8E6 A0A1W4WPT7 A0A2H8TFE7 J9JKN0 A0A2S2NV02 E0VAP2 J9HSC3 A0A1B0CRR2 A0A1W7R8C8 A0A1B0D1S5 J3JUA4 A0A069DQD9 A0A224XI03 A0A182QF31 A0A182VRS1 A0A182LSI1 A0A182NLI5 A0A146M7A4 A0A0A9Z8Q7 A0A0M9A8L6 A0A182XV05 A0A182RPC3 A0A182WSW9 A0A182V3V9 A0A182LA81 A0A182TSI2 A0A182HLS3 A0A2S2R4Y7 Q7PY55 A0A182SMS8 A0A1Y1KU79 A0A2A3ECA0 W5JFH6 A0A182NZC9 A0A1B0GNT7 A0A182FNL1 A0A1W4X0U4 A0A0L7QSV8 T1HQ12 A0A182KA05 A0A182JE34 A0A067R8Z3 A0A0T6B6V3 A0A1W4X358 A0A1Y1MAW1 A0A2P8Z2R9 A0A1W4XDZ2 A0A2J7QR46 A0A2P8Y4B5 D6WT10 A0A140KPR8 D6WG91 A0A2P8ZLP3 R4FLI9 Q6RUR3 A0A2J7QR36 A0A1W4X3A0 A0A084W563 A0A1Y1N5U0 A0A151WLU8 A0A151XEI7 A0A023F7A3 A0A067R8A9 A0A1Y1N8U2 V5GSR5 A0A224XI56 A0A1Y1MYW2 A0A1Y1N8K6 A0A154PE73 A0A023FB52 A0A224XI78
PDB
2GDZ
E-value=3.16381e-39,
Score=404
Ontologies
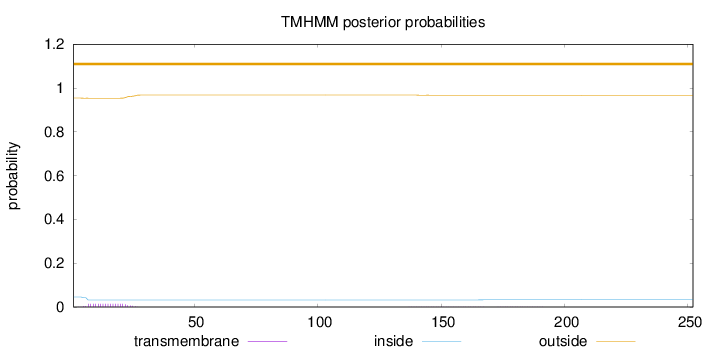
Topology
Length:
252
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29032
Exp number, first 60 AAs:
0.26025
Total prob of N-in:
0.04589
outside
1 - 252
Population Genetic Test Statistics
Pi
176.503139
Theta
162.875197
Tajima's D
0.683101
CLR
0.012638
CSRT
0.564121793910304
Interpretation
Uncertain
