Gene
KWMTBOMO06476 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011619
Annotation
PREDICTED:_protein_LSM14_homolog_B_isoform_X3_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.542
Sequence
CDS
ATGAGTTCTGGAATGCCTGAATTAGGCTCCAAAATAAGCCTAATATCGAAAGCTGACATTCGTTACGAAGGCCGACTTTTTACTGTCGATCCTCAGGAATGTACTATCGCCTTGGCTAGCGTGCGTTCATTTGGCACGGAGGATCGAGAGACTCAATATCCGGTAGCACCTCAAAGCCAAGTATATGACTACATTCTGTTCCGTGGTTCTGACATCAAGGATATCAGGGTGCTAAATAATGTTCCTTCAATGCCCAATGATCCTGCCATTATGCAGATGTCTGTACCCCCAACTCTGGGTACTGGACCAGCTCCTGGGCAGTATGTGGGGCAGTTTAACCATCCAGTTGTTGGACAAACTCCATATCCTCAGTATCATCCAATGGCTGGCTTTCCAAGTGGAGTTCACAACCAGCTCAACAAGACGAGCTCAATGCTTGACCTCATTGGTGGAGGATCCCAACAAAATGCATCTCGGTCAGGTACACCTGCTGTCGGTGTTCATAGGAAAAGTCCAACAGCTGACCAAAGCATTCAGGTTGGTACAGGCGCTGGCAGCTCAGGTCAACAACGGGATAGCAGACGCGGGAGCAACAACACTGCACAGCAACAACAGCAGCAGCAGCAGCAACAACAAAAGGGCCAGCAACGCAATCGTACCAACTCACGCAACCGACAACGTTTTCCGAGTGGCACGAATCAGCAGCAACCCCATCAACCACTACAACAACAACAACAACAACAACAACCTCAGCAACAGAATATGCAACACCAACCTAGACCTCAGCATCACAATCATCAAGGGTTTTTTGATGGACAAAACTTTAATCGTGGAGGATGGCGAGGCAGGCCACGTGGACGTGGCCGTGGTGGATTCATACCTCGAAACAAGAACACACTCAAGTTCGACAATGATTATGATTTTGAACAGGCCAATACTGAATTTGAAGAACTGAGGAATCAACTTGCCAAAACTAAGATCAGTGAAGGAGATGTAAAAGTTGAGGGAACAGTAGAAGTGGACAAGAAAGACGATTCTGGCAACGAAACGGGTGCCGGCGAACCTGAAGTCGAGGAAGATTCGTCTTTCACTGGCTATGACAAGACCAAGAGCTTCTTTGACAACATATCCTGTGAGGCTGTTGAGAGGTCAAAGGGGAGGTCTCAGCGGACGGATTGGCGTACGGAGCGTAAGCTGAACTCAGAGACGTTTGGCGTAGCGTCGGCGCGGCGCGGGGCCTGGCGCCCTCGCGCGTCGTGGCGTCATCCCGCGCACCACAACCAACACAATCAACATAACCAACAGTACGCGCATCACCACTACCAGCAGCAACATGCCTGGCGCTCCAACCGCGGTGCGCGCTCTCGGCCCTCCACCAGCACCGCCGCGCCGGAACAGTCTGCACCAGCTAAGTAG
Protein
MSSGMPELGSKISLISKADIRYEGRLFTVDPQECTIALASVRSFGTEDRETQYPVAPQSQVYDYILFRGSDIKDIRVLNNVPSMPNDPAIMQMSVPPTLGTGPAPGQYVGQFNHPVVGQTPYPQYHPMAGFPSGVHNQLNKTSSMLDLIGGGSQQNASRSGTPAVGVHRKSPTADQSIQVGTGAGSSGQQRDSRRGSNNTAQQQQQQQQQQQKGQQRNRTNSRNRQRFPSGTNQQQPHQPLQQQQQQQQPQQQNMQHQPRPQHHNHQGFFDGQNFNRGGWRGRPRGRGRGGFIPRNKNTLKFDNDYDFEQANTEFEELRNQLAKTKISEGDVKVEGTVEVDKKDDSGNETGAGEPEVEEDSSFTGYDKTKSFFDNISCEAVERSKGRSQRTDWRTERKLNSETFGVASARRGAWRPRASWRHPAHHNQHNQHNQQYAHHHYQQQHAWRSNRGARSRPSTSTAAPEQSAPAK
Summary
Uniprot
ProteinModelPortal
PDB
2VXE
E-value=1.69091e-35,
Score=375
Ontologies
GO
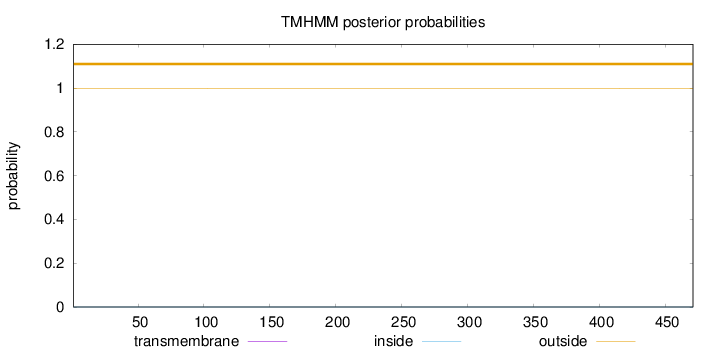
Topology
Length:
471
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00044
outside
1 - 471
Population Genetic Test Statistics
Pi
240.70647
Theta
206.444218
Tajima's D
0.447262
CLR
0.113078
CSRT
0.503624818759062
Interpretation
Uncertain
