Gene
KWMTBOMO06469
Pre Gene Modal
BGIBMGA011617
Annotation
PREDICTED:_lipase_member_H-like_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.024 Mitochondrial Reliability : 1.067
Sequence
CDS
ATGAAGCAATTAGCCAAAGACCCGAACATGGATTTCAGTAAGCGAACAGCTGTATACGTCGGAGGGTATATGGACAGTCCGAGTTATCCAGTTGCAAGTCTATTGGCCCCACAGTATCGACGCCAAGGATATAATGTTCTGCTGTTGGACACAAATAAGTTTACGGTCATGGAGTATCCTTTGGCTGTCCGCTACATAAGAACCGCTGCCCGACACACAGCAGAGATGCTAGTCGACCTAATAAAGCACGGTCTGGATCCAAAAAAACTAGACCTGATTGGCCTTAGTTTGGGCGCGCACACAGCCAGCTTCATCGCGAAACGCTTCAAGGAACTAACCGGTGATACCATATCGAGACTCACCGGGTTAGATCCAGCAGGACCGTGCTTCAGGAATTTGGGCCCCGATCAGCGACTGGACTCGTCTGATGCGGACTTCGTGGACGTCGTCGATACGAATATAGACGAGTTTGGTATGGCGGCTCCGATTGGCCACGTAAATTTCTACGTTAACGGAGGCGAGTACCAACCTGGCGATATTCTTTGGATGCCGTGCAACGCCTTCTGCAGTCATCTAAGGTCTTACACTGTTTGGTTGGCGGCACTGCAGTATCCGACCTCGTTTATAGCAATACAGTGCGATTCTGTGCAAGAGGCGAGAAATAAAAATTGCTACGATAGAATACCTTTGGTCACGAACGTTGTCGGTTTGGGTACGGACAAGAGTAAAACTGGTGTATTTTACTTAGCAACGCACAATAATTATCCGTATTATTTAGGAGAAAAGGGACTGAAGAAGGAATACGAATATTTCCAGTCTCATATGGAAGAAATTACCAATAATGATAGTATGAAAATCTGA
Protein
MKQLAKDPNMDFSKRTAVYVGGYMDSPSYPVASLLAPQYRRQGYNVLLLDTNKFTVMEYPLAVRYIRTAARHTAEMLVDLIKHGLDPKKLDLIGLSLGAHTASFIAKRFKELTGDTISRLTGLDPAGPCFRNLGPDQRLDSSDADFVDVVDTNIDEFGMAAPIGHVNFYVNGGEYQPGDILWMPCNAFCSHLRSYTVWLAALQYPTSFIAIQCDSVQEARNKNCYDRIPLVTNVVGLGTDKSKTGVFYLATHNNYPYYLGEKGLKKEYEYFQSHMEEITNNDSMKI
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
H9JQ12
A0A194Q1D6
A0A0N1II42
A0A2H1WK50
A0A2W1B572
A0A212FBR8
+ More
H9JQQ8 A0A194QBX2 A0A2W1BNT7 A0A194QUH6 A0A2H1WL24 H9JHY8 A0A0L7LHY6 A0A212FPW6 A0A3S2LBT4 A0A0L7L1G4 A0A194QBZ0 A0A191XQW7 A0A2A4JBH9 A0A2H1VJ13 A0A0N1IGL3 A0A3S2PK48 A0A2A4JEV7 A0A3S2P6E3 A0A2H1VGU5 A0A2A4JXD4 A0A2W1BL36 A0A0L7KV46 A0A194QZ66 A0A212EY15 H9JXD2 A0A194PEP3 H9IUE6 A0A212EJL3 I6TFN4 A0A3S2NZZ7 A0A194RQL7 A0A3S2L3L9 A0A0L7KP71 A0A194PSE8 H9JWY8 A0A2W1BWG2 A0A2A4JGD9 A0A2H1X3Q9 A0A194PCA8 A0A2H1WL84 A0A0L7L8R4 A0A194QNS4 A0A2H1W795 A0A212FKA5 A0A2A4J690 A0A194QC74 A0A2H1W7F1 A0A2A4JYV7 A0A2H1WNR8 A0A220K8L9 A0A2W1BXY3 A0A0N0PB11 H9JFT5 A0A2H1W7Q1 A0A2W1BV63 A0A2A4J1M9 A0A2A4J5Z0 S4RQL2 D6WBV6 A0A2A4IT83 A0A3P4MXK8 A0A3B3ZZW1 F7DME2 A0A1S3GQV1 A0A0N1PJG6 A0A3P8WEC3 A0A1B6E9S5 A0A232EHJ7 A0A1I8MKM1 V9KDH1 A0A3Q3D8S5 A0A1A8ABA8 A0A0L7LP71 A0A3Q1CJ28 A0A1U7Q4B7 A0A1A8RSL5 A0A2Y9JC22 A0A3P8S6J0 A0A1A8CJJ6 A0A1A8HX65 A0A1A8DHY5 A0A1A8K8Z5 A0A1A8UNF0 A0A3P8S465
H9JQQ8 A0A194QBX2 A0A2W1BNT7 A0A194QUH6 A0A2H1WL24 H9JHY8 A0A0L7LHY6 A0A212FPW6 A0A3S2LBT4 A0A0L7L1G4 A0A194QBZ0 A0A191XQW7 A0A2A4JBH9 A0A2H1VJ13 A0A0N1IGL3 A0A3S2PK48 A0A2A4JEV7 A0A3S2P6E3 A0A2H1VGU5 A0A2A4JXD4 A0A2W1BL36 A0A0L7KV46 A0A194QZ66 A0A212EY15 H9JXD2 A0A194PEP3 H9IUE6 A0A212EJL3 I6TFN4 A0A3S2NZZ7 A0A194RQL7 A0A3S2L3L9 A0A0L7KP71 A0A194PSE8 H9JWY8 A0A2W1BWG2 A0A2A4JGD9 A0A2H1X3Q9 A0A194PCA8 A0A2H1WL84 A0A0L7L8R4 A0A194QNS4 A0A2H1W795 A0A212FKA5 A0A2A4J690 A0A194QC74 A0A2H1W7F1 A0A2A4JYV7 A0A2H1WNR8 A0A220K8L9 A0A2W1BXY3 A0A0N0PB11 H9JFT5 A0A2H1W7Q1 A0A2W1BV63 A0A2A4J1M9 A0A2A4J5Z0 S4RQL2 D6WBV6 A0A2A4IT83 A0A3P4MXK8 A0A3B3ZZW1 F7DME2 A0A1S3GQV1 A0A0N1PJG6 A0A3P8WEC3 A0A1B6E9S5 A0A232EHJ7 A0A1I8MKM1 V9KDH1 A0A3Q3D8S5 A0A1A8ABA8 A0A0L7LP71 A0A3Q1CJ28 A0A1U7Q4B7 A0A1A8RSL5 A0A2Y9JC22 A0A3P8S6J0 A0A1A8CJJ6 A0A1A8HX65 A0A1A8DHY5 A0A1A8K8Z5 A0A1A8UNF0 A0A3P8S465
Pubmed
EMBL
BABH01012334
KQ459590
KPI97210.1
KQ460391
KPJ15413.1
ODYU01009136
+ More
SOQ53346.1 KZ150534 PZC70591.1 AGBW02009288 OWR51181.1 KQ459193 KPJ03053.1 KZ149947 PZC76719.1 KQ461154 KPJ08630.1 ODYU01009344 SOQ53707.1 BABH01035306 JTDY01001036 KOB75067.1 AGBW02000859 OWR55743.1 RSAL01000052 RVE50204.1 JTDY01003560 KOB69343.1 KPJ03068.1 KU360126 ANJ42864.1 NWSH01002209 PCG68904.1 ODYU01002823 SOQ40786.1 KQ460781 KPJ12226.1 RSAL01000010 RVE53725.1 NWSH01001665 PCG70481.1 RSAL01000291 RVE42897.1 ODYU01002500 SOQ40069.1 NWSH01000400 PCG76675.1 KZ149983 PZC75749.1 JTDY01005546 KOB66899.1 KQ460947 KPJ10569.1 AGBW02011630 OWR46389.1 BABH01035864 KQ459606 KPI91164.1 BABH01001181 AGBW02014435 OWR41693.1 JQ814807 AFM73623.1 RSAL01000012 RVE53468.1 KQ459833 KPJ19782.1 RSAL01000184 RVE44850.1 JTDY01007594 KOB65092.1 KQ459601 KPI94050.1 BABH01041781 KZ149926 PZC77547.1 NWSH01001670 PCG70462.1 ODYU01013233 SOQ59929.1 KPI90911.1 ODYU01009427 SOQ53845.1 JTDY01002277 KOB71750.1 KQ461196 KPJ06625.1 ODYU01006796 SOQ48975.1 AGBW02008082 OWR54177.1 NWSH01003103 PCG66943.1 KPJ03024.1 SOQ48977.1 NWSH01000413 PCG76562.1 ODYU01009953 SOQ54715.1 MF319718 ASJ26432.1 PZC77546.1 KQ461072 KPJ09835.1 BABH01001653 SOQ48976.1 PZC77545.1 NWSH01003717 PCG65891.1 PCG66944.1 KQ971315 EEZ97864.2 NWSH01007523 PCG62931.1 CYRY02015547 VCW85512.1 KQ460240 KPJ16392.1 GEDC01002625 JAS34673.1 NNAY01004496 OXU17813.1 JW863333 AFO95850.1 HADY01013306 SBP51791.1 JTDY01000429 KOB77220.1 HAEI01009602 SBS08234.1 HADZ01014994 SBP78935.1 HAED01002857 SBQ88715.1 HAEA01005480 SBQ33960.1 HAEE01008773 SBR28823.1 HAEJ01009148 SBS49605.1
SOQ53346.1 KZ150534 PZC70591.1 AGBW02009288 OWR51181.1 KQ459193 KPJ03053.1 KZ149947 PZC76719.1 KQ461154 KPJ08630.1 ODYU01009344 SOQ53707.1 BABH01035306 JTDY01001036 KOB75067.1 AGBW02000859 OWR55743.1 RSAL01000052 RVE50204.1 JTDY01003560 KOB69343.1 KPJ03068.1 KU360126 ANJ42864.1 NWSH01002209 PCG68904.1 ODYU01002823 SOQ40786.1 KQ460781 KPJ12226.1 RSAL01000010 RVE53725.1 NWSH01001665 PCG70481.1 RSAL01000291 RVE42897.1 ODYU01002500 SOQ40069.1 NWSH01000400 PCG76675.1 KZ149983 PZC75749.1 JTDY01005546 KOB66899.1 KQ460947 KPJ10569.1 AGBW02011630 OWR46389.1 BABH01035864 KQ459606 KPI91164.1 BABH01001181 AGBW02014435 OWR41693.1 JQ814807 AFM73623.1 RSAL01000012 RVE53468.1 KQ459833 KPJ19782.1 RSAL01000184 RVE44850.1 JTDY01007594 KOB65092.1 KQ459601 KPI94050.1 BABH01041781 KZ149926 PZC77547.1 NWSH01001670 PCG70462.1 ODYU01013233 SOQ59929.1 KPI90911.1 ODYU01009427 SOQ53845.1 JTDY01002277 KOB71750.1 KQ461196 KPJ06625.1 ODYU01006796 SOQ48975.1 AGBW02008082 OWR54177.1 NWSH01003103 PCG66943.1 KPJ03024.1 SOQ48977.1 NWSH01000413 PCG76562.1 ODYU01009953 SOQ54715.1 MF319718 ASJ26432.1 PZC77546.1 KQ461072 KPJ09835.1 BABH01001653 SOQ48976.1 PZC77545.1 NWSH01003717 PCG65891.1 PCG66944.1 KQ971315 EEZ97864.2 NWSH01007523 PCG62931.1 CYRY02015547 VCW85512.1 KQ460240 KPJ16392.1 GEDC01002625 JAS34673.1 NNAY01004496 OXU17813.1 JW863333 AFO95850.1 HADY01013306 SBP51791.1 JTDY01000429 KOB77220.1 HAEI01009602 SBS08234.1 HADZ01014994 SBP78935.1 HAED01002857 SBQ88715.1 HAEA01005480 SBQ33960.1 HAEE01008773 SBR28823.1 HAEJ01009148 SBS49605.1
Proteomes
Pfam
PF00151 Lipase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JQ12
A0A194Q1D6
A0A0N1II42
A0A2H1WK50
A0A2W1B572
A0A212FBR8
+ More
H9JQQ8 A0A194QBX2 A0A2W1BNT7 A0A194QUH6 A0A2H1WL24 H9JHY8 A0A0L7LHY6 A0A212FPW6 A0A3S2LBT4 A0A0L7L1G4 A0A194QBZ0 A0A191XQW7 A0A2A4JBH9 A0A2H1VJ13 A0A0N1IGL3 A0A3S2PK48 A0A2A4JEV7 A0A3S2P6E3 A0A2H1VGU5 A0A2A4JXD4 A0A2W1BL36 A0A0L7KV46 A0A194QZ66 A0A212EY15 H9JXD2 A0A194PEP3 H9IUE6 A0A212EJL3 I6TFN4 A0A3S2NZZ7 A0A194RQL7 A0A3S2L3L9 A0A0L7KP71 A0A194PSE8 H9JWY8 A0A2W1BWG2 A0A2A4JGD9 A0A2H1X3Q9 A0A194PCA8 A0A2H1WL84 A0A0L7L8R4 A0A194QNS4 A0A2H1W795 A0A212FKA5 A0A2A4J690 A0A194QC74 A0A2H1W7F1 A0A2A4JYV7 A0A2H1WNR8 A0A220K8L9 A0A2W1BXY3 A0A0N0PB11 H9JFT5 A0A2H1W7Q1 A0A2W1BV63 A0A2A4J1M9 A0A2A4J5Z0 S4RQL2 D6WBV6 A0A2A4IT83 A0A3P4MXK8 A0A3B3ZZW1 F7DME2 A0A1S3GQV1 A0A0N1PJG6 A0A3P8WEC3 A0A1B6E9S5 A0A232EHJ7 A0A1I8MKM1 V9KDH1 A0A3Q3D8S5 A0A1A8ABA8 A0A0L7LP71 A0A3Q1CJ28 A0A1U7Q4B7 A0A1A8RSL5 A0A2Y9JC22 A0A3P8S6J0 A0A1A8CJJ6 A0A1A8HX65 A0A1A8DHY5 A0A1A8K8Z5 A0A1A8UNF0 A0A3P8S465
H9JQQ8 A0A194QBX2 A0A2W1BNT7 A0A194QUH6 A0A2H1WL24 H9JHY8 A0A0L7LHY6 A0A212FPW6 A0A3S2LBT4 A0A0L7L1G4 A0A194QBZ0 A0A191XQW7 A0A2A4JBH9 A0A2H1VJ13 A0A0N1IGL3 A0A3S2PK48 A0A2A4JEV7 A0A3S2P6E3 A0A2H1VGU5 A0A2A4JXD4 A0A2W1BL36 A0A0L7KV46 A0A194QZ66 A0A212EY15 H9JXD2 A0A194PEP3 H9IUE6 A0A212EJL3 I6TFN4 A0A3S2NZZ7 A0A194RQL7 A0A3S2L3L9 A0A0L7KP71 A0A194PSE8 H9JWY8 A0A2W1BWG2 A0A2A4JGD9 A0A2H1X3Q9 A0A194PCA8 A0A2H1WL84 A0A0L7L8R4 A0A194QNS4 A0A2H1W795 A0A212FKA5 A0A2A4J690 A0A194QC74 A0A2H1W7F1 A0A2A4JYV7 A0A2H1WNR8 A0A220K8L9 A0A2W1BXY3 A0A0N0PB11 H9JFT5 A0A2H1W7Q1 A0A2W1BV63 A0A2A4J1M9 A0A2A4J5Z0 S4RQL2 D6WBV6 A0A2A4IT83 A0A3P4MXK8 A0A3B3ZZW1 F7DME2 A0A1S3GQV1 A0A0N1PJG6 A0A3P8WEC3 A0A1B6E9S5 A0A232EHJ7 A0A1I8MKM1 V9KDH1 A0A3Q3D8S5 A0A1A8ABA8 A0A0L7LP71 A0A3Q1CJ28 A0A1U7Q4B7 A0A1A8RSL5 A0A2Y9JC22 A0A3P8S6J0 A0A1A8CJJ6 A0A1A8HX65 A0A1A8DHY5 A0A1A8K8Z5 A0A1A8UNF0 A0A3P8S465
PDB
1W52
E-value=7.87019e-21,
Score=246
Ontologies
GO
PANTHER
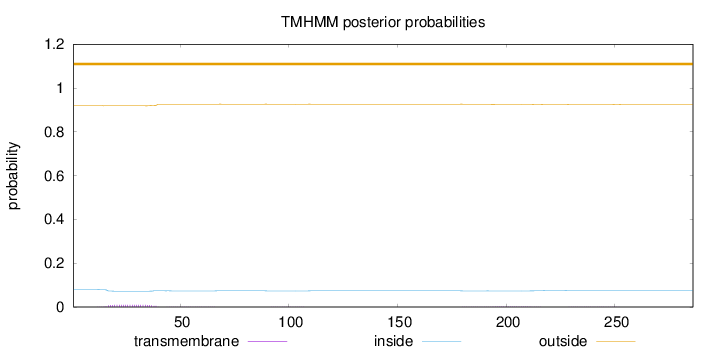
Topology
Subcellular location
Secreted
Length:
286
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.272120000000001
Exp number, first 60 AAs:
0.21713
Total prob of N-in:
0.07931
outside
1 - 286
Population Genetic Test Statistics
Pi
195.682598
Theta
146.062424
Tajima's D
1.217349
CLR
0.000004
CSRT
0.719314034298285
Interpretation
Uncertain
