Gene
KWMTBOMO06464
Pre Gene Modal
BGIBMGA011865
Annotation
PREDICTED:_uncharacterized_protein_LOC106121342_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 2.387
Sequence
CDS
ATGGCGCACAAATTAAAGAACAACCATTATGTAGCCACGCATCGTTCTGGCGTAGATTCGCCGTCATCCGGAGGATATTTGCACAGAGCATCGTCTAGAGATGAAAACGACAGCCACAGAGCACCTTCGGAGAGGACGCTGTCAGAGTACACGACAATGGACGAAAGAACCCGATCGCCGTCTGGCAATGACCACCGGAACGGCAGGCGGCACCAGCAAACGAACGGCTCGCCTCGGGCCGACTCCGCGTCCGATGTCTACGTCACTAGCGCTGCCTACAGACCTCCTTCGGAAATCAGTCGGCAGTCCCGTGCTCCTCGCAGCGTTTATTCCTACCGCAGCGCCGTAGCGCCATCTGTCGCGTCTACACACAGATCCAAAGTTTCTAGGAAGGCAGGAGTGAAAGTAGACGCGATGGCAGCGCCGAACCCGTTCTGTCCGAACGTGAAGGGCATGTGCTGTCTGATGCTGCTCCTCAACCTGGGGCTCATCCTTGTCACACTGGGCTTCGTTATCGTGCTCCAGTTCTTCGAGCCGCTGTTTGTTTGGATCTTGGGTATAGTCTTCTTGATATTCGGCTTCATAACTCTAGTGGGAAGTCTCGTGTACTGCGTAGTTTTGTGCCGAGAGAACCCCTATCCTCGATACCCGGATGACTTCCACTGGACCCACCATTGGTCTAAAACAATAGGGCCGTCGGAAATACATTACAATGCCAGCGAGAAGCAGTACCGGCAGAATGGACACAATAGGTACAATGGCAAATATTCGGACAGGGAAAGTGCAAAAGGCTACTCTGATAGAGAAAGCAAGCTGAGTAGGTACTGA
Protein
MAHKLKNNHYVATHRSGVDSPSSGGYLHRASSRDENDSHRAPSERTLSEYTTMDERTRSPSGNDHRNGRRHQQTNGSPRADSASDVYVTSAAYRPPSEISRQSRAPRSVYSYRSAVAPSVASTHRSKVSRKAGVKVDAMAAPNPFCPNVKGMCCLMLLLNLGLILVTLGFVIVLQFFEPLFVWILGIVFLIFGFITLVGSLVYCVVLCRENPYPRYPDDFHWTHHWSKTIGPSEIHYNASEKQYRQNGHNRYNGKYSDRESAKGYSDRESKLSRY
Summary
Uniprot
A0A2W1BYQ1
A0A194PV59
A0A3S2M899
A0A0N1IPG4
A0A1E1WIN5
A0A2P8YWA0
+ More
A0A1Y1LIB2 A0A1Y1LKF6 A0A067RKT8 A0A084WDF7 A0A182JJU0 N6ST54 D6WF62 A0A182NJH6 A0A182K8Q6 A0A182PIG3 A0A182V992 A0A182U6Q3 A0A182XMG6 Q7Q6J3 A0A182WL78 A0A182QZJ2 A0A182MHH5 A0A182R2A9 W5JQZ9 A0A182YJA9 A0A182GXE7 A0A182FDW3 Q16XD7 A0A2M3Z346 A0A2M3Z3F7 A0A2M4AE00 A0A2M4BUU4 A0A182I296 A0A1W4XD73 T1PM24 X1WJN0 A0A0L0CH36 A0A1I8QCK7 A0A1A9VE99 A0A1A9ZF20 A0A1B0G331 A0A1A9YFI2 A0A1B0BRC3 A0A1A9W7M4 A0A1Q3FQ11 A0A1B6DYU1 A0A182KQ33 A0A1B6EPL2 A0A2S2PNK1 A0A1Q3FQE0 A0A1W4UGT3 B4PK48 A0A1B6GVI3 A0A2S2PVC7 B4HAS8 Q29E83 Q8SYN3 B3NFG1 A0A3B0JPY7 B4HV39 A0A0J9RQR5 Q9VS39 B3M4Z1 A0A1B6I2Q3 A0A1J1J3K4 B4LC90 B4QKB4 A0A0M4EEX5 B0XJ00 B4L959 A0A2M4AL84 B4MLD2 A0A2M4AK02 B4J1Y9 A0A1L8D8R5 A0A336LV31 A0A0K8WEN3 A0A151IAB2 A0A034WSL8 A0A0T6BDB3 A0A0J7KYU7 E2AU90 A0A195BE50 F4WBM7 A0A151X5W8 A0A158NRI2 A0A195DJG7 A0A195FU20 A0A1B6EE30 E2BMU1 A0A1B6H1Z9 T1HCJ5 A0A0A9YNK3 A0A1L8D997
A0A1Y1LIB2 A0A1Y1LKF6 A0A067RKT8 A0A084WDF7 A0A182JJU0 N6ST54 D6WF62 A0A182NJH6 A0A182K8Q6 A0A182PIG3 A0A182V992 A0A182U6Q3 A0A182XMG6 Q7Q6J3 A0A182WL78 A0A182QZJ2 A0A182MHH5 A0A182R2A9 W5JQZ9 A0A182YJA9 A0A182GXE7 A0A182FDW3 Q16XD7 A0A2M3Z346 A0A2M3Z3F7 A0A2M4AE00 A0A2M4BUU4 A0A182I296 A0A1W4XD73 T1PM24 X1WJN0 A0A0L0CH36 A0A1I8QCK7 A0A1A9VE99 A0A1A9ZF20 A0A1B0G331 A0A1A9YFI2 A0A1B0BRC3 A0A1A9W7M4 A0A1Q3FQ11 A0A1B6DYU1 A0A182KQ33 A0A1B6EPL2 A0A2S2PNK1 A0A1Q3FQE0 A0A1W4UGT3 B4PK48 A0A1B6GVI3 A0A2S2PVC7 B4HAS8 Q29E83 Q8SYN3 B3NFG1 A0A3B0JPY7 B4HV39 A0A0J9RQR5 Q9VS39 B3M4Z1 A0A1B6I2Q3 A0A1J1J3K4 B4LC90 B4QKB4 A0A0M4EEX5 B0XJ00 B4L959 A0A2M4AL84 B4MLD2 A0A2M4AK02 B4J1Y9 A0A1L8D8R5 A0A336LV31 A0A0K8WEN3 A0A151IAB2 A0A034WSL8 A0A0T6BDB3 A0A0J7KYU7 E2AU90 A0A195BE50 F4WBM7 A0A151X5W8 A0A158NRI2 A0A195DJG7 A0A195FU20 A0A1B6EE30 E2BMU1 A0A1B6H1Z9 T1HCJ5 A0A0A9YNK3 A0A1L8D997
Pubmed
28756777
26354079
29403074
28004739
24845553
24438588
+ More
23537049 18362917 19820115 12364791 14747013 17210077 20920257 23761445 25244985 26483478 17510324 25315136 26108605 20966253 17994087 17550304 15632085 23185243 18057021 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 20798317 21719571 21347285 25401762 26823975
23537049 18362917 19820115 12364791 14747013 17210077 20920257 23761445 25244985 26483478 17510324 25315136 26108605 20966253 17994087 17550304 15632085 23185243 18057021 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 20798317 21719571 21347285 25401762 26823975
EMBL
KZ149896
PZC78775.1
KQ459590
KPI97207.1
RSAL01000010
RVE53728.1
+ More
KQ460391 KPJ15410.1 GDQN01004297 JAT86757.1 PYGN01000323 PSN48522.1 GEZM01054802 JAV73392.1 GEZM01054803 JAV73391.1 KK852567 KDR21200.1 ATLV01022999 KE525339 KFB48251.1 APGK01057337 KB741280 KB631604 ENN70874.1 ERL84594.1 KQ971327 EEZ99837.1 AAAB01008960 EAA10794.2 AXCN02000445 AXCM01002279 ADMH02000425 ETN66546.1 JXUM01095110 JXUM01095111 KQ564173 KXJ72629.1 CH477541 EAT39275.1 GGFM01002193 MBW22944.1 GGFM01002310 MBW23061.1 GGFK01005695 MBW39016.1 GGFJ01007643 MBW56784.1 APCN01000134 KA649200 KA649837 AFP64466.1 ABLF02038748 ABLF02038754 ABLF02038760 ABLF02038761 JRES01000493 KNC30804.1 CCAG010009844 CCAG010009845 CCAG010009846 JXJN01019040 GFDL01005356 JAV29689.1 GEDC01006455 JAS30843.1 GECZ01030016 JAS39753.1 GGMR01017797 MBY30416.1 GFDL01005339 JAV29706.1 CM000159 EDW93728.1 KRK01327.1 KRK01328.1 KRK01329.1 GECZ01003340 JAS66429.1 GGMS01000166 MBY69369.1 CH479244 EDW37710.1 CH379070 EAL30179.1 KRT08699.1 AY071427 BT056263 AAL49049.1 ACL68710.1 CH954178 EDV50503.1 KQS43328.1 OUUW01000002 SPP77530.1 CH480817 EDW50810.1 CM002912 KMY98044.1 KMY98045.1 KMY98046.1 KMY98047.1 AE014296 BT133242 AAF50590.1 AFB18665.1 CH902618 EDV40565.1 KPU78627.1 GECU01026502 JAS81204.1 CVRI01000068 CRL06959.1 CRL06960.1 CH940647 EDW68735.1 CM000363 EDX09537.1 CP012525 ALC44408.1 DS233417 EDS29882.1 CH933816 EDW17234.1 GGFK01008235 MBW41556.1 CH963847 EDW73390.1 GGFK01007756 MBW41077.1 CH916366 EDV95914.1 GFDF01011242 JAV02842.1 UFQS01000127 UFQT01000127 SSX00169.1 SSX20549.1 GDHF01024164 GDHF01002693 JAI28150.1 JAI49621.1 KQ978229 KYM96221.1 GAKP01002204 JAC56748.1 LJIG01001645 KRT85295.1 LBMM01001958 KMQ95473.1 GL442815 EFN62947.1 KQ976511 KYM82445.1 GL888066 EGI68305.1 KQ982498 KYQ55680.1 ADTU01024046 KQ980824 KYN12629.1 KQ981280 KYN43384.1 GEDC01001127 JAS36171.1 GL449347 EFN82976.1 GECZ01001078 JAS68691.1 ACPB03004527 GBHO01010926 GBRD01006785 GDHC01013588 JAG32678.1 JAG59036.1 JAQ05041.1 GFDF01011056 JAV03028.1
KQ460391 KPJ15410.1 GDQN01004297 JAT86757.1 PYGN01000323 PSN48522.1 GEZM01054802 JAV73392.1 GEZM01054803 JAV73391.1 KK852567 KDR21200.1 ATLV01022999 KE525339 KFB48251.1 APGK01057337 KB741280 KB631604 ENN70874.1 ERL84594.1 KQ971327 EEZ99837.1 AAAB01008960 EAA10794.2 AXCN02000445 AXCM01002279 ADMH02000425 ETN66546.1 JXUM01095110 JXUM01095111 KQ564173 KXJ72629.1 CH477541 EAT39275.1 GGFM01002193 MBW22944.1 GGFM01002310 MBW23061.1 GGFK01005695 MBW39016.1 GGFJ01007643 MBW56784.1 APCN01000134 KA649200 KA649837 AFP64466.1 ABLF02038748 ABLF02038754 ABLF02038760 ABLF02038761 JRES01000493 KNC30804.1 CCAG010009844 CCAG010009845 CCAG010009846 JXJN01019040 GFDL01005356 JAV29689.1 GEDC01006455 JAS30843.1 GECZ01030016 JAS39753.1 GGMR01017797 MBY30416.1 GFDL01005339 JAV29706.1 CM000159 EDW93728.1 KRK01327.1 KRK01328.1 KRK01329.1 GECZ01003340 JAS66429.1 GGMS01000166 MBY69369.1 CH479244 EDW37710.1 CH379070 EAL30179.1 KRT08699.1 AY071427 BT056263 AAL49049.1 ACL68710.1 CH954178 EDV50503.1 KQS43328.1 OUUW01000002 SPP77530.1 CH480817 EDW50810.1 CM002912 KMY98044.1 KMY98045.1 KMY98046.1 KMY98047.1 AE014296 BT133242 AAF50590.1 AFB18665.1 CH902618 EDV40565.1 KPU78627.1 GECU01026502 JAS81204.1 CVRI01000068 CRL06959.1 CRL06960.1 CH940647 EDW68735.1 CM000363 EDX09537.1 CP012525 ALC44408.1 DS233417 EDS29882.1 CH933816 EDW17234.1 GGFK01008235 MBW41556.1 CH963847 EDW73390.1 GGFK01007756 MBW41077.1 CH916366 EDV95914.1 GFDF01011242 JAV02842.1 UFQS01000127 UFQT01000127 SSX00169.1 SSX20549.1 GDHF01024164 GDHF01002693 JAI28150.1 JAI49621.1 KQ978229 KYM96221.1 GAKP01002204 JAC56748.1 LJIG01001645 KRT85295.1 LBMM01001958 KMQ95473.1 GL442815 EFN62947.1 KQ976511 KYM82445.1 GL888066 EGI68305.1 KQ982498 KYQ55680.1 ADTU01024046 KQ980824 KYN12629.1 KQ981280 KYN43384.1 GEDC01001127 JAS36171.1 GL449347 EFN82976.1 GECZ01001078 JAS68691.1 ACPB03004527 GBHO01010926 GBRD01006785 GDHC01013588 JAG32678.1 JAG59036.1 JAQ05041.1 GFDF01011056 JAV03028.1
Proteomes
UP000053268
UP000283053
UP000053240
UP000245037
UP000027135
UP000030765
+ More
UP000075880 UP000019118 UP000030742 UP000007266 UP000075884 UP000075881 UP000075885 UP000075903 UP000075902 UP000076407 UP000007062 UP000075920 UP000075886 UP000075883 UP000075900 UP000000673 UP000076408 UP000069940 UP000249989 UP000069272 UP000008820 UP000075840 UP000192223 UP000095301 UP000007819 UP000037069 UP000095300 UP000078200 UP000092445 UP000092444 UP000092443 UP000092460 UP000091820 UP000075882 UP000192221 UP000002282 UP000008744 UP000001819 UP000008711 UP000268350 UP000001292 UP000000803 UP000007801 UP000183832 UP000008792 UP000000304 UP000092553 UP000002320 UP000009192 UP000007798 UP000001070 UP000078542 UP000036403 UP000000311 UP000078540 UP000007755 UP000075809 UP000005205 UP000078492 UP000078541 UP000008237 UP000015103
UP000075880 UP000019118 UP000030742 UP000007266 UP000075884 UP000075881 UP000075885 UP000075903 UP000075902 UP000076407 UP000007062 UP000075920 UP000075886 UP000075883 UP000075900 UP000000673 UP000076408 UP000069940 UP000249989 UP000069272 UP000008820 UP000075840 UP000192223 UP000095301 UP000007819 UP000037069 UP000095300 UP000078200 UP000092445 UP000092444 UP000092443 UP000092460 UP000091820 UP000075882 UP000192221 UP000002282 UP000008744 UP000001819 UP000008711 UP000268350 UP000001292 UP000000803 UP000007801 UP000183832 UP000008792 UP000000304 UP000092553 UP000002320 UP000009192 UP000007798 UP000001070 UP000078542 UP000036403 UP000000311 UP000078540 UP000007755 UP000075809 UP000005205 UP000078492 UP000078541 UP000008237 UP000015103
ProteinModelPortal
A0A2W1BYQ1
A0A194PV59
A0A3S2M899
A0A0N1IPG4
A0A1E1WIN5
A0A2P8YWA0
+ More
A0A1Y1LIB2 A0A1Y1LKF6 A0A067RKT8 A0A084WDF7 A0A182JJU0 N6ST54 D6WF62 A0A182NJH6 A0A182K8Q6 A0A182PIG3 A0A182V992 A0A182U6Q3 A0A182XMG6 Q7Q6J3 A0A182WL78 A0A182QZJ2 A0A182MHH5 A0A182R2A9 W5JQZ9 A0A182YJA9 A0A182GXE7 A0A182FDW3 Q16XD7 A0A2M3Z346 A0A2M3Z3F7 A0A2M4AE00 A0A2M4BUU4 A0A182I296 A0A1W4XD73 T1PM24 X1WJN0 A0A0L0CH36 A0A1I8QCK7 A0A1A9VE99 A0A1A9ZF20 A0A1B0G331 A0A1A9YFI2 A0A1B0BRC3 A0A1A9W7M4 A0A1Q3FQ11 A0A1B6DYU1 A0A182KQ33 A0A1B6EPL2 A0A2S2PNK1 A0A1Q3FQE0 A0A1W4UGT3 B4PK48 A0A1B6GVI3 A0A2S2PVC7 B4HAS8 Q29E83 Q8SYN3 B3NFG1 A0A3B0JPY7 B4HV39 A0A0J9RQR5 Q9VS39 B3M4Z1 A0A1B6I2Q3 A0A1J1J3K4 B4LC90 B4QKB4 A0A0M4EEX5 B0XJ00 B4L959 A0A2M4AL84 B4MLD2 A0A2M4AK02 B4J1Y9 A0A1L8D8R5 A0A336LV31 A0A0K8WEN3 A0A151IAB2 A0A034WSL8 A0A0T6BDB3 A0A0J7KYU7 E2AU90 A0A195BE50 F4WBM7 A0A151X5W8 A0A158NRI2 A0A195DJG7 A0A195FU20 A0A1B6EE30 E2BMU1 A0A1B6H1Z9 T1HCJ5 A0A0A9YNK3 A0A1L8D997
A0A1Y1LIB2 A0A1Y1LKF6 A0A067RKT8 A0A084WDF7 A0A182JJU0 N6ST54 D6WF62 A0A182NJH6 A0A182K8Q6 A0A182PIG3 A0A182V992 A0A182U6Q3 A0A182XMG6 Q7Q6J3 A0A182WL78 A0A182QZJ2 A0A182MHH5 A0A182R2A9 W5JQZ9 A0A182YJA9 A0A182GXE7 A0A182FDW3 Q16XD7 A0A2M3Z346 A0A2M3Z3F7 A0A2M4AE00 A0A2M4BUU4 A0A182I296 A0A1W4XD73 T1PM24 X1WJN0 A0A0L0CH36 A0A1I8QCK7 A0A1A9VE99 A0A1A9ZF20 A0A1B0G331 A0A1A9YFI2 A0A1B0BRC3 A0A1A9W7M4 A0A1Q3FQ11 A0A1B6DYU1 A0A182KQ33 A0A1B6EPL2 A0A2S2PNK1 A0A1Q3FQE0 A0A1W4UGT3 B4PK48 A0A1B6GVI3 A0A2S2PVC7 B4HAS8 Q29E83 Q8SYN3 B3NFG1 A0A3B0JPY7 B4HV39 A0A0J9RQR5 Q9VS39 B3M4Z1 A0A1B6I2Q3 A0A1J1J3K4 B4LC90 B4QKB4 A0A0M4EEX5 B0XJ00 B4L959 A0A2M4AL84 B4MLD2 A0A2M4AK02 B4J1Y9 A0A1L8D8R5 A0A336LV31 A0A0K8WEN3 A0A151IAB2 A0A034WSL8 A0A0T6BDB3 A0A0J7KYU7 E2AU90 A0A195BE50 F4WBM7 A0A151X5W8 A0A158NRI2 A0A195DJG7 A0A195FU20 A0A1B6EE30 E2BMU1 A0A1B6H1Z9 T1HCJ5 A0A0A9YNK3 A0A1L8D997
Ontologies
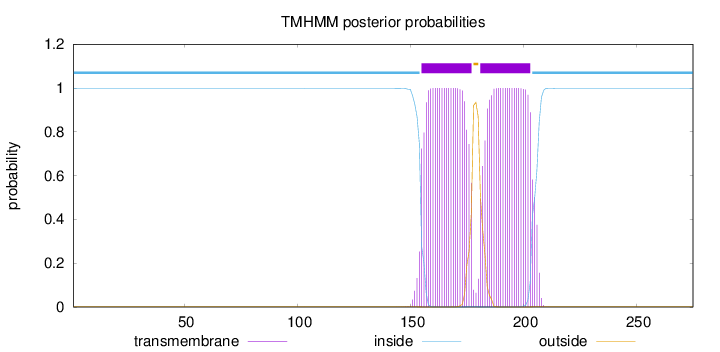
Topology
Length:
275
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.48008
Exp number, first 60 AAs:
0
Total prob of N-in:
0.99872
inside
1 - 154
TMhelix
155 - 177
outside
178 - 180
TMhelix
181 - 203
inside
204 - 275
Population Genetic Test Statistics
Pi
260.018121
Theta
18.166913
Tajima's D
1.213591
CLR
0.536421
CSRT
0.725963701814909
Interpretation
Uncertain
