Pre Gene Modal
BGIBMGA011867
Annotation
Transmembrane_protein_223_[Papilio_machaon]
Location in the cell
Mitochondrial Reliability : 2.749
Sequence
CDS
ATGTACACCCTTCGTTCCGTGAGATATTTAGTTCTCCACAAAGGAGGCAAGCATGTTTCATTGGTCACTTACGCTCCATTTGGTCGCAATCGATCCATGAAAGTTCCCATTGAAGATATTTGCTGTCAGGAAAGAAGGACAGTGGCAAAAGTACAGCTACCTCTCAAAGTGAGAAATAGAATGATGCATTATATGCTGGATATGAGGGGAGAGTTTAAAAACCCTTTGCTGTTTGATTGTACCGCAGGCTTACTCAGGAAATGGGGGAAAAGTTAG
Protein
MYTLRSVRYLVLHKGGKHVSLVTYAPFGRNRSMKVPIEDICCQERRTVAKVQLPLKVRNRMMHYMLDMRGEFKNPLLFDCTAGLLRKWGKS
Summary
Uniprot
H9JQR1
A0A2A4KB59
A0A0N1PJP7
A0A2W1C3N6
A0A2H1VFB4
A0A194Q1D0
+ More
A0A1E1WBW5 S4PI65 A0A0L7LQR2 A0A212FJN4 D6WF50 A0A3B0J461 A0A1Y1K5Q3 A0A336KP43 A0A3R7PQ11 A0A1A9YC14 A0A1B0BHK5 A0A0A1WMT4 A0A0K8WLI2 A0A034WKR2 A0A1A9VY05 A0A1A9ZX61 W8AKR5 Q28XJ8 B4GG94 B4LKP3 B4KQ84 B4MNY6 A0A0T6AWL0 A0A1I8PCE4 B4J7T8 A0A224XXX3 T1HD35 A0A1W4VPZ5 A0A1I8MBX8 Q7PQ06 A0A1J1HRC9 A0A084W2G0 A0A0L0CBT6 A0A182SYG2 A0A0P4VN95 A0A182XJB9 A0A182V198 A0A182LBF4 A7UT85 A0A182HP42 A0A182UA44 B4P835 B3NN52 A0A182YNQ2 A0A034WI67 B4HN01 B4QAM9 A0A182WAD3 A0A182K2B9 A0A182P596 A0A182M8W2 A0A0A9YW34 Q7JW07 B3MIK3 A0A2R7WLP1 A0A182N866 A0A2J7QSS9 A0A182RTS0 A0A0M4EHZ9 A0A069DQ37 A0A1B0DQB3 A0A182FE48 A0A2M3ZD21 W5JMD2 A0A2P8Z8C2 A0A2M4AXI0 A0A2M4BYH4 A0A2M4AY56 A0A3R7NXC2 A0A2M4AY38 A0A182J6C0 A0A1A9WYI8 A0A182GZU8 A0A1Q3FHE5 A0A067RNB7 A0A1B6CAU7 A0A1L8D9E1 B0WW13 A0A1L8D9P7 B0XHI6 Q0IEZ9 A0A1B6FNX2 A0A1B6HE11 A0A1B6LX19 A0A170Y2T8 A0A0P5HLS3 A0A0P5QR31 A0A0N8AZ06 N6UMA1 A0A0P6F592 A0A0P5HDV7 A0A0P6HQS9 A0A0K8TLN9 A0A0P6BSK6
A0A1E1WBW5 S4PI65 A0A0L7LQR2 A0A212FJN4 D6WF50 A0A3B0J461 A0A1Y1K5Q3 A0A336KP43 A0A3R7PQ11 A0A1A9YC14 A0A1B0BHK5 A0A0A1WMT4 A0A0K8WLI2 A0A034WKR2 A0A1A9VY05 A0A1A9ZX61 W8AKR5 Q28XJ8 B4GG94 B4LKP3 B4KQ84 B4MNY6 A0A0T6AWL0 A0A1I8PCE4 B4J7T8 A0A224XXX3 T1HD35 A0A1W4VPZ5 A0A1I8MBX8 Q7PQ06 A0A1J1HRC9 A0A084W2G0 A0A0L0CBT6 A0A182SYG2 A0A0P4VN95 A0A182XJB9 A0A182V198 A0A182LBF4 A7UT85 A0A182HP42 A0A182UA44 B4P835 B3NN52 A0A182YNQ2 A0A034WI67 B4HN01 B4QAM9 A0A182WAD3 A0A182K2B9 A0A182P596 A0A182M8W2 A0A0A9YW34 Q7JW07 B3MIK3 A0A2R7WLP1 A0A182N866 A0A2J7QSS9 A0A182RTS0 A0A0M4EHZ9 A0A069DQ37 A0A1B0DQB3 A0A182FE48 A0A2M3ZD21 W5JMD2 A0A2P8Z8C2 A0A2M4AXI0 A0A2M4BYH4 A0A2M4AY56 A0A3R7NXC2 A0A2M4AY38 A0A182J6C0 A0A1A9WYI8 A0A182GZU8 A0A1Q3FHE5 A0A067RNB7 A0A1B6CAU7 A0A1L8D9E1 B0WW13 A0A1L8D9P7 B0XHI6 Q0IEZ9 A0A1B6FNX2 A0A1B6HE11 A0A1B6LX19 A0A170Y2T8 A0A0P5HLS3 A0A0P5QR31 A0A0N8AZ06 N6UMA1 A0A0P6F592 A0A0P5HDV7 A0A0P6HQS9 A0A0K8TLN9 A0A0P6BSK6
Pubmed
19121390
26354079
28756777
23622113
26227816
22118469
+ More
18362917 19820115 28004739 25830018 25348373 24495485 15632085 17994087 25315136 12364791 14747013 17210077 24438588 26108605 27129103 20966253 17550304 25244985 22936249 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 20920257 23761445 29403074 26483478 24845553 17510324 23537049 26369729
18362917 19820115 28004739 25830018 25348373 24495485 15632085 17994087 25315136 12364791 14747013 17210077 24438588 26108605 27129103 20966253 17550304 25244985 22936249 25401762 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26334808 20920257 23761445 29403074 26483478 24845553 17510324 23537049 26369729
EMBL
BABH01012346
NWSH01000006
PCG80980.1
KQ460391
KPJ15408.1
KZ149896
+ More
PZC78773.1 ODYU01002280 SOQ39550.1 KQ459590 KPI97205.1 GDQN01006588 JAT84466.1 GAIX01005520 JAA87040.1 JTDY01000313 KOB77739.1 AGBW02008251 OWR53942.1 KQ971327 EEZ99844.1 OUUW01000001 SPP74152.1 GEZM01091432 JAV56849.1 UFQS01000749 UFQT01000749 SSX06576.1 SSX26923.1 QCYY01001300 ROT79046.1 JXJN01014402 GBXI01013963 JAD00329.1 GDHF01000313 JAI52001.1 GAKP01004579 JAC54373.1 GAMC01020123 GAMC01020121 GAMC01020119 JAB86432.1 CM000071 EAL26318.1 CH479183 EDW35514.1 CH940648 EDW61766.1 CH933808 EDW09212.1 CH963848 EDW73825.1 LJIG01022685 KRT79261.1 CH916367 EDW01144.1 GFTR01003016 JAW13410.1 ACPB03012555 AAAB01008900 EAA09462.5 CVRI01000010 CRK89030.1 ATLV01019615 KE525275 KFB44404.1 JRES01000637 KNC29702.1 GDKW01003372 JAI53223.1 EDO64090.1 APCN01002909 CM000158 EDW90082.1 CH954179 EDV56572.1 GAKP01004578 JAC54374.1 CH480816 EDW47305.1 CM000362 CM002911 EDX06534.1 KMY92852.1 AXCM01002472 GBHO01006392 GBRD01006213 GDHC01001775 JAG37212.1 JAG59608.1 JAQ16854.1 AE013599 AY118499 AAF58754.1 AAM49868.1 CH902619 EDV38079.1 KK855050 PTY20524.1 NEVH01011228 PNF31625.1 CP012524 ALC40630.1 GBGD01003152 JAC85737.1 AJVK01019007 GGFM01005660 MBW26411.1 ADMH02000690 ETN65306.1 PYGN01000150 PSN52737.1 GGFK01012159 MBW45480.1 GGFJ01008995 MBW58136.1 GGFK01012197 MBW45518.1 QCYY01002569 ROT69357.1 GGFK01012177 MBW45498.1 JXUM01002838 KQ560145 KXJ84186.1 GFDL01008055 JAV26990.1 KK852567 KDR21199.1 GEDC01026849 JAS10449.1 GFDF01011006 JAV03078.1 DS232136 EDS35845.1 GFDF01011007 JAV03077.1 DS233161 EDS28415.1 CH477440 EAT40882.1 GECZ01018570 GECZ01017915 JAS51199.1 JAS51854.1 GECU01034865 JAS72841.1 GEBQ01011731 JAT28246.1 GEMB01003724 JAR99496.1 GDIQ01231265 JAK20460.1 GDIQ01120548 JAL31178.1 GDIQ01229011 JAK22714.1 APGK01026530 KB740635 ENN79842.1 GDIQ01067824 JAN26913.1 GDIQ01231264 JAK20461.1 GDIQ01040085 JAN54652.1 GDAI01002349 JAI15254.1 GDIP01011792 JAM91923.1
PZC78773.1 ODYU01002280 SOQ39550.1 KQ459590 KPI97205.1 GDQN01006588 JAT84466.1 GAIX01005520 JAA87040.1 JTDY01000313 KOB77739.1 AGBW02008251 OWR53942.1 KQ971327 EEZ99844.1 OUUW01000001 SPP74152.1 GEZM01091432 JAV56849.1 UFQS01000749 UFQT01000749 SSX06576.1 SSX26923.1 QCYY01001300 ROT79046.1 JXJN01014402 GBXI01013963 JAD00329.1 GDHF01000313 JAI52001.1 GAKP01004579 JAC54373.1 GAMC01020123 GAMC01020121 GAMC01020119 JAB86432.1 CM000071 EAL26318.1 CH479183 EDW35514.1 CH940648 EDW61766.1 CH933808 EDW09212.1 CH963848 EDW73825.1 LJIG01022685 KRT79261.1 CH916367 EDW01144.1 GFTR01003016 JAW13410.1 ACPB03012555 AAAB01008900 EAA09462.5 CVRI01000010 CRK89030.1 ATLV01019615 KE525275 KFB44404.1 JRES01000637 KNC29702.1 GDKW01003372 JAI53223.1 EDO64090.1 APCN01002909 CM000158 EDW90082.1 CH954179 EDV56572.1 GAKP01004578 JAC54374.1 CH480816 EDW47305.1 CM000362 CM002911 EDX06534.1 KMY92852.1 AXCM01002472 GBHO01006392 GBRD01006213 GDHC01001775 JAG37212.1 JAG59608.1 JAQ16854.1 AE013599 AY118499 AAF58754.1 AAM49868.1 CH902619 EDV38079.1 KK855050 PTY20524.1 NEVH01011228 PNF31625.1 CP012524 ALC40630.1 GBGD01003152 JAC85737.1 AJVK01019007 GGFM01005660 MBW26411.1 ADMH02000690 ETN65306.1 PYGN01000150 PSN52737.1 GGFK01012159 MBW45480.1 GGFJ01008995 MBW58136.1 GGFK01012197 MBW45518.1 QCYY01002569 ROT69357.1 GGFK01012177 MBW45498.1 JXUM01002838 KQ560145 KXJ84186.1 GFDL01008055 JAV26990.1 KK852567 KDR21199.1 GEDC01026849 JAS10449.1 GFDF01011006 JAV03078.1 DS232136 EDS35845.1 GFDF01011007 JAV03077.1 DS233161 EDS28415.1 CH477440 EAT40882.1 GECZ01018570 GECZ01017915 JAS51199.1 JAS51854.1 GECU01034865 JAS72841.1 GEBQ01011731 JAT28246.1 GEMB01003724 JAR99496.1 GDIQ01231265 JAK20460.1 GDIQ01120548 JAL31178.1 GDIQ01229011 JAK22714.1 APGK01026530 KB740635 ENN79842.1 GDIQ01067824 JAN26913.1 GDIQ01231264 JAK20461.1 GDIQ01040085 JAN54652.1 GDAI01002349 JAI15254.1 GDIP01011792 JAM91923.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000007266 UP000268350 UP000283509 UP000092443 UP000092460 UP000078200 UP000092445 UP000001819 UP000008744 UP000008792 UP000009192 UP000007798 UP000095300 UP000001070 UP000015103 UP000192221 UP000095301 UP000007062 UP000183832 UP000030765 UP000037069 UP000075901 UP000076407 UP000075903 UP000075882 UP000075840 UP000075902 UP000002282 UP000008711 UP000076408 UP000001292 UP000000304 UP000075920 UP000075881 UP000075885 UP000075883 UP000000803 UP000007801 UP000075884 UP000235965 UP000075900 UP000092553 UP000092462 UP000069272 UP000000673 UP000245037 UP000075880 UP000091820 UP000069940 UP000249989 UP000027135 UP000002320 UP000008820 UP000019118
UP000007266 UP000268350 UP000283509 UP000092443 UP000092460 UP000078200 UP000092445 UP000001819 UP000008744 UP000008792 UP000009192 UP000007798 UP000095300 UP000001070 UP000015103 UP000192221 UP000095301 UP000007062 UP000183832 UP000030765 UP000037069 UP000075901 UP000076407 UP000075903 UP000075882 UP000075840 UP000075902 UP000002282 UP000008711 UP000076408 UP000001292 UP000000304 UP000075920 UP000075881 UP000075885 UP000075883 UP000000803 UP000007801 UP000075884 UP000235965 UP000075900 UP000092553 UP000092462 UP000069272 UP000000673 UP000245037 UP000075880 UP000091820 UP000069940 UP000249989 UP000027135 UP000002320 UP000008820 UP000019118
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JQR1
A0A2A4KB59
A0A0N1PJP7
A0A2W1C3N6
A0A2H1VFB4
A0A194Q1D0
+ More
A0A1E1WBW5 S4PI65 A0A0L7LQR2 A0A212FJN4 D6WF50 A0A3B0J461 A0A1Y1K5Q3 A0A336KP43 A0A3R7PQ11 A0A1A9YC14 A0A1B0BHK5 A0A0A1WMT4 A0A0K8WLI2 A0A034WKR2 A0A1A9VY05 A0A1A9ZX61 W8AKR5 Q28XJ8 B4GG94 B4LKP3 B4KQ84 B4MNY6 A0A0T6AWL0 A0A1I8PCE4 B4J7T8 A0A224XXX3 T1HD35 A0A1W4VPZ5 A0A1I8MBX8 Q7PQ06 A0A1J1HRC9 A0A084W2G0 A0A0L0CBT6 A0A182SYG2 A0A0P4VN95 A0A182XJB9 A0A182V198 A0A182LBF4 A7UT85 A0A182HP42 A0A182UA44 B4P835 B3NN52 A0A182YNQ2 A0A034WI67 B4HN01 B4QAM9 A0A182WAD3 A0A182K2B9 A0A182P596 A0A182M8W2 A0A0A9YW34 Q7JW07 B3MIK3 A0A2R7WLP1 A0A182N866 A0A2J7QSS9 A0A182RTS0 A0A0M4EHZ9 A0A069DQ37 A0A1B0DQB3 A0A182FE48 A0A2M3ZD21 W5JMD2 A0A2P8Z8C2 A0A2M4AXI0 A0A2M4BYH4 A0A2M4AY56 A0A3R7NXC2 A0A2M4AY38 A0A182J6C0 A0A1A9WYI8 A0A182GZU8 A0A1Q3FHE5 A0A067RNB7 A0A1B6CAU7 A0A1L8D9E1 B0WW13 A0A1L8D9P7 B0XHI6 Q0IEZ9 A0A1B6FNX2 A0A1B6HE11 A0A1B6LX19 A0A170Y2T8 A0A0P5HLS3 A0A0P5QR31 A0A0N8AZ06 N6UMA1 A0A0P6F592 A0A0P5HDV7 A0A0P6HQS9 A0A0K8TLN9 A0A0P6BSK6
A0A1E1WBW5 S4PI65 A0A0L7LQR2 A0A212FJN4 D6WF50 A0A3B0J461 A0A1Y1K5Q3 A0A336KP43 A0A3R7PQ11 A0A1A9YC14 A0A1B0BHK5 A0A0A1WMT4 A0A0K8WLI2 A0A034WKR2 A0A1A9VY05 A0A1A9ZX61 W8AKR5 Q28XJ8 B4GG94 B4LKP3 B4KQ84 B4MNY6 A0A0T6AWL0 A0A1I8PCE4 B4J7T8 A0A224XXX3 T1HD35 A0A1W4VPZ5 A0A1I8MBX8 Q7PQ06 A0A1J1HRC9 A0A084W2G0 A0A0L0CBT6 A0A182SYG2 A0A0P4VN95 A0A182XJB9 A0A182V198 A0A182LBF4 A7UT85 A0A182HP42 A0A182UA44 B4P835 B3NN52 A0A182YNQ2 A0A034WI67 B4HN01 B4QAM9 A0A182WAD3 A0A182K2B9 A0A182P596 A0A182M8W2 A0A0A9YW34 Q7JW07 B3MIK3 A0A2R7WLP1 A0A182N866 A0A2J7QSS9 A0A182RTS0 A0A0M4EHZ9 A0A069DQ37 A0A1B0DQB3 A0A182FE48 A0A2M3ZD21 W5JMD2 A0A2P8Z8C2 A0A2M4AXI0 A0A2M4BYH4 A0A2M4AY56 A0A3R7NXC2 A0A2M4AY38 A0A182J6C0 A0A1A9WYI8 A0A182GZU8 A0A1Q3FHE5 A0A067RNB7 A0A1B6CAU7 A0A1L8D9E1 B0WW13 A0A1L8D9P7 B0XHI6 Q0IEZ9 A0A1B6FNX2 A0A1B6HE11 A0A1B6LX19 A0A170Y2T8 A0A0P5HLS3 A0A0P5QR31 A0A0N8AZ06 N6UMA1 A0A0P6F592 A0A0P5HDV7 A0A0P6HQS9 A0A0K8TLN9 A0A0P6BSK6
Ontologies
PANTHER
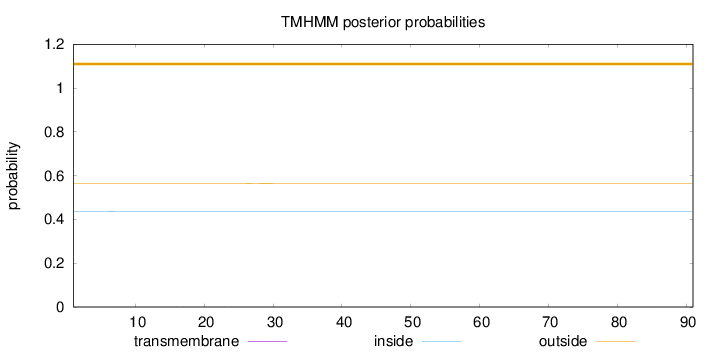
Topology
Length:
91
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02092
Exp number, first 60 AAs:
0.01783
Total prob of N-in:
0.43572
outside
1 - 91
Population Genetic Test Statistics
Pi
211.967087
Theta
172.677173
Tajima's D
0.715755
CLR
0.045988
CSRT
0.579771011449428
Interpretation
Uncertain
