Gene
KWMTBOMO06456
Pre Gene Modal
BGIBMGA011869
Annotation
PREDICTED:_uncharacterized_protein_LOC101743192_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.635
Sequence
CDS
ATGATTCTACCGCGTGTTGCGCAACAAAATATTTTGAACTGTAGGTCAATAAGTGGTTCGCGATGTTGTTATCAAAACTTGGAAAGTAAATACTCGGAAAGTCAAAAAGTGAAAATACTAAATGTAATTAATGATGATTCTCAAACATTGTCAAGGTTTGATATAGCAAAGTCAAGATTAAAAAAGTTTAAACAATGGAAAACTAGCAATGGTCAAGTGAAGACCCTATCTGATTTACCTTTAATAGAAGGTTTCACTGACAAGACAGCTAAGAAACTCTGTGACAGTATATTAAATGGACCCACTGAAGAAGTTGAACAAATATCAAATAAAATAAAAGGTCAAATATTACATCCAAATTTGAAAGAAAGTACTATTAAAGACTGTAAAACAGTATTGACAGTATATATATCTGTTAATTCAGTTTGTTGGACGCTTATAAACAAAAATGATTATGAAGTTGTGGAGTGGCAGTATTATGGTATTGATTACCCCGAAGGAAAGAAGATTCAGATCACAGATATATTTGATATTGCATGGCGTATAACGCAGCGGTTACCGGTCGCTGACATTTATGTGATGAAAGCTGAAGCTACCACTTTGCGAGCGGCCGGTAGCGATCCGAACAATCCTAAAGTATTGGCTGTAAATCTTCAGAAAGCTCAAATGGTCTCTATGATTGTTGCATTGATTAACGCACGAGGACACAGAAGTGACGATGACGATGGTAAATTGAAACAGATTGTTTATTTTTTGAGGCCCACATTACCCTACAGGCTACACGGAACTTTAGTTGGAAACGAAAGAGTGTCGACTGATCAAACCGTCGAAATGTTACTTCAGGAGAGCAGTGGTAAATCAACAGGCAACAAACACGTCTACGTGCCGGAGGAAGGGAAAAACATGTTCAGGACACAGAACGAACTACAGAAGGACATGTTGGGACATTGTCTGTTACTTAGTTTGACTTTTATGGATTTGTGTGTTTACAAGAACAAGGAGCGCATCGCGAAATTAATTAAGAGGACTTCTTGA
Protein
MILPRVAQQNILNCRSISGSRCCYQNLESKYSESQKVKILNVINDDSQTLSRFDIAKSRLKKFKQWKTSNGQVKTLSDLPLIEGFTDKTAKKLCDSILNGPTEEVEQISNKIKGQILHPNLKESTIKDCKTVLTVYISVNSVCWTLINKNDYEVVEWQYYGIDYPEGKKIQITDIFDIAWRITQRLPVADIYVMKAEATTLRAAGSDPNNPKVLAVNLQKAQMVSMIVALINARGHRSDDDDGKLKQIVYFLRPTLPYRLHGTLVGNERVSTDQTVEMLLQESSGKSTGNKHVYVPEEGKNMFRTQNELQKDMLGHCLLLSLTFMDLCVYKNKERIAKLIKRTS
Summary
Uniprot
H9JQR3
A0A2W1BUX4
A0A2A4K9L3
A0A2H1WVD6
A0A0N1PH52
A0A194Q1C5
+ More
A0A212ELU7 A0A2P8XE23 A0A1J1I3U8 A0A1W4URC5 A0A067QYD0 A0A2J7QGZ2 A0A1Y1KF72 B4N5G2 B4KYJ1 D6WF54 B4HC18 Q16YC8 A0A1S4FK08 Q29F15 A0A023EQB2 B4J3Q1 A0A0K8W3L1 A0A0A1XGT1 A0A182IQW6 A0A034W8H5 B4LDY5 A0A1I8QB38 A0A0M4F0T2 A0A084WHJ9 W8CBT7 Q9VNR9 A0A182QT18 A0A3B0JUA2 B4QLA0 B4IB37 A0A182WG71 N6TSB9 A0A182MYN4 B3MB80 B4PIP6 A0A182RTI9 A0A182MS58 A0A182P6I1 A0A0J7KIR1 Q5TN70 A0A182UH13 A0A182SV23 A0A182WZL7 A0A182V4A6 A0A182HQ63 A0A182JZ05 A0A1B0B6X3 A0A182Y1P5 A0A2M4BS69 A0A182FW68 A0A2M4AUR5 A0A0T6B6M6 A0A2M4AUX8 E9IUR0 E2ACB6 A0A0L0C8C7 A0A1B0EXZ7 A0A1I8MS43 B0WFZ5 A0A1A9WD82 A0A026WZM7 A0A3L8DPS2 A0A2M4BQU4 A0A0Q5UL26 A0A1A9VFA8 A0A1B0A2D3 W5JIQ5 A0A195DXY4 A0A1B0FB80 F4WZN9 A0A195B6X2 A0A158NH77 A0A1B6BW40 A0A151XCU8 A0A195FFI4 A0A1A9Y0Y5 A0A1B6ERF1 A0A1B6HEL0 A0A232FKF6 A0A3B1JWN3 A0A293MU96 A0A2M4AXM3 A0A1B6M7L5 A0A060X5C3 A0A1S3L5U4 A0A3Q1HIP3 A0A1A8IR40 A0A2R5LMS0
A0A212ELU7 A0A2P8XE23 A0A1J1I3U8 A0A1W4URC5 A0A067QYD0 A0A2J7QGZ2 A0A1Y1KF72 B4N5G2 B4KYJ1 D6WF54 B4HC18 Q16YC8 A0A1S4FK08 Q29F15 A0A023EQB2 B4J3Q1 A0A0K8W3L1 A0A0A1XGT1 A0A182IQW6 A0A034W8H5 B4LDY5 A0A1I8QB38 A0A0M4F0T2 A0A084WHJ9 W8CBT7 Q9VNR9 A0A182QT18 A0A3B0JUA2 B4QLA0 B4IB37 A0A182WG71 N6TSB9 A0A182MYN4 B3MB80 B4PIP6 A0A182RTI9 A0A182MS58 A0A182P6I1 A0A0J7KIR1 Q5TN70 A0A182UH13 A0A182SV23 A0A182WZL7 A0A182V4A6 A0A182HQ63 A0A182JZ05 A0A1B0B6X3 A0A182Y1P5 A0A2M4BS69 A0A182FW68 A0A2M4AUR5 A0A0T6B6M6 A0A2M4AUX8 E9IUR0 E2ACB6 A0A0L0C8C7 A0A1B0EXZ7 A0A1I8MS43 B0WFZ5 A0A1A9WD82 A0A026WZM7 A0A3L8DPS2 A0A2M4BQU4 A0A0Q5UL26 A0A1A9VFA8 A0A1B0A2D3 W5JIQ5 A0A195DXY4 A0A1B0FB80 F4WZN9 A0A195B6X2 A0A158NH77 A0A1B6BW40 A0A151XCU8 A0A195FFI4 A0A1A9Y0Y5 A0A1B6ERF1 A0A1B6HEL0 A0A232FKF6 A0A3B1JWN3 A0A293MU96 A0A2M4AXM3 A0A1B6M7L5 A0A060X5C3 A0A1S3L5U4 A0A3Q1HIP3 A0A1A8IR40 A0A2R5LMS0
Pubmed
19121390
28756777
26354079
22118469
29403074
24845553
+ More
28004739 17994087 18362917 19820115 17510324 15632085 24945155 25830018 25348373 24438588 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 23537049 17550304 12364791 14747013 17210077 25244985 21282665 20798317 26108605 25315136 24508170 30249741 20920257 23761445 21719571 21347285 28648823 25329095 24755649
28004739 17994087 18362917 19820115 17510324 15632085 24945155 25830018 25348373 24438588 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 23537049 17550304 12364791 14747013 17210077 25244985 21282665 20798317 26108605 25315136 24508170 30249741 20920257 23761445 21719571 21347285 28648823 25329095 24755649
EMBL
BABH01012358
BABH01012359
BABH01012360
KZ149896
PZC78769.1
NWSH01000006
+ More
PCG80985.1 ODYU01010947 SOQ56374.1 KQ460391 KPJ15403.1 KQ459590 KPI97200.1 AGBW02013978 OWR42464.1 PYGN01002565 PSN30256.1 CVRI01000040 CRK95015.1 KK852879 KDR14455.1 NEVH01014359 PNF27856.1 GEZM01087213 JAV58840.1 CH964101 EDW79601.1 CH933809 EDW18802.2 KQ971327 EEZ99841.2 CH479275 EDW40049.1 CH477518 EAT39642.1 CH379070 EAL29894.2 GAPW01002325 JAC11273.1 CH916366 EDV97282.1 GDHF01006566 JAI45748.1 GBXI01004125 JAD10167.1 GAKP01007081 JAC51871.1 CH940647 EDW70028.1 CP012525 ALC44800.1 ATLV01023844 KE525347 KFB49693.1 GAMC01004531 JAC02025.1 AE014296 AY071634 AAF51853.1 AAL49256.1 AXCN02001036 OUUW01000002 SPP76301.1 CM000363 CM002912 EDX11518.1 KMZ01218.1 CH480826 EDW44500.1 APGK01021195 KB740293 ENN80953.1 CH902618 EDV41381.1 CM000159 EDW95554.1 AXCM01002364 LBMM01006992 KMQ90137.1 AAAB01008986 EAL38690.3 APCN01003297 JXJN01009258 GGFJ01006497 MBW55638.1 GGFK01011216 MBW44537.1 LJIG01009476 KRT83004.1 GGFK01011250 MBW44571.1 GL766012 EFZ15714.1 GL438491 EFN68906.1 JRES01000763 KNC28482.1 AJVK01033360 DS231921 EDS26530.1 KK107063 EZA61181.1 QOIP01000005 RLU22424.1 GGFJ01006314 MBW55455.1 CH954178 KQS44434.1 ADMH02001285 ETN63173.1 KQ980107 KYN17686.1 CCAG010017825 GL888480 EGI60314.1 KQ976574 KYM80246.1 ADTU01015205 GEDC01031786 JAS05512.1 KQ982294 KYQ58202.1 KQ981625 KYN39116.1 GECZ01029321 JAS40448.1 GECU01034588 JAS73118.1 NNAY01000084 OXU31142.1 GFWV01018803 MAA43531.1 GGFK01012213 MBW45534.1 GEBQ01008050 JAT31927.1 FR904998 CDQ74763.1 HAED01013346 SBQ99791.1 GGLE01006710 MBY10836.1
PCG80985.1 ODYU01010947 SOQ56374.1 KQ460391 KPJ15403.1 KQ459590 KPI97200.1 AGBW02013978 OWR42464.1 PYGN01002565 PSN30256.1 CVRI01000040 CRK95015.1 KK852879 KDR14455.1 NEVH01014359 PNF27856.1 GEZM01087213 JAV58840.1 CH964101 EDW79601.1 CH933809 EDW18802.2 KQ971327 EEZ99841.2 CH479275 EDW40049.1 CH477518 EAT39642.1 CH379070 EAL29894.2 GAPW01002325 JAC11273.1 CH916366 EDV97282.1 GDHF01006566 JAI45748.1 GBXI01004125 JAD10167.1 GAKP01007081 JAC51871.1 CH940647 EDW70028.1 CP012525 ALC44800.1 ATLV01023844 KE525347 KFB49693.1 GAMC01004531 JAC02025.1 AE014296 AY071634 AAF51853.1 AAL49256.1 AXCN02001036 OUUW01000002 SPP76301.1 CM000363 CM002912 EDX11518.1 KMZ01218.1 CH480826 EDW44500.1 APGK01021195 KB740293 ENN80953.1 CH902618 EDV41381.1 CM000159 EDW95554.1 AXCM01002364 LBMM01006992 KMQ90137.1 AAAB01008986 EAL38690.3 APCN01003297 JXJN01009258 GGFJ01006497 MBW55638.1 GGFK01011216 MBW44537.1 LJIG01009476 KRT83004.1 GGFK01011250 MBW44571.1 GL766012 EFZ15714.1 GL438491 EFN68906.1 JRES01000763 KNC28482.1 AJVK01033360 DS231921 EDS26530.1 KK107063 EZA61181.1 QOIP01000005 RLU22424.1 GGFJ01006314 MBW55455.1 CH954178 KQS44434.1 ADMH02001285 ETN63173.1 KQ980107 KYN17686.1 CCAG010017825 GL888480 EGI60314.1 KQ976574 KYM80246.1 ADTU01015205 GEDC01031786 JAS05512.1 KQ982294 KYQ58202.1 KQ981625 KYN39116.1 GECZ01029321 JAS40448.1 GECU01034588 JAS73118.1 NNAY01000084 OXU31142.1 GFWV01018803 MAA43531.1 GGFK01012213 MBW45534.1 GEBQ01008050 JAT31927.1 FR904998 CDQ74763.1 HAED01013346 SBQ99791.1 GGLE01006710 MBY10836.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000245037
+ More
UP000183832 UP000192221 UP000027135 UP000235965 UP000007798 UP000009192 UP000007266 UP000008744 UP000008820 UP000001819 UP000001070 UP000075880 UP000008792 UP000095300 UP000092553 UP000030765 UP000000803 UP000075886 UP000268350 UP000000304 UP000001292 UP000075920 UP000019118 UP000075884 UP000007801 UP000002282 UP000075900 UP000075883 UP000075885 UP000036403 UP000007062 UP000075902 UP000075901 UP000076407 UP000075903 UP000075840 UP000075881 UP000092460 UP000076408 UP000069272 UP000000311 UP000037069 UP000092462 UP000095301 UP000002320 UP000091820 UP000053097 UP000279307 UP000008711 UP000078200 UP000092445 UP000000673 UP000078492 UP000092444 UP000007755 UP000078540 UP000005205 UP000075809 UP000078541 UP000092443 UP000215335 UP000018467 UP000193380 UP000087266 UP000265040
UP000183832 UP000192221 UP000027135 UP000235965 UP000007798 UP000009192 UP000007266 UP000008744 UP000008820 UP000001819 UP000001070 UP000075880 UP000008792 UP000095300 UP000092553 UP000030765 UP000000803 UP000075886 UP000268350 UP000000304 UP000001292 UP000075920 UP000019118 UP000075884 UP000007801 UP000002282 UP000075900 UP000075883 UP000075885 UP000036403 UP000007062 UP000075902 UP000075901 UP000076407 UP000075903 UP000075840 UP000075881 UP000092460 UP000076408 UP000069272 UP000000311 UP000037069 UP000092462 UP000095301 UP000002320 UP000091820 UP000053097 UP000279307 UP000008711 UP000078200 UP000092445 UP000000673 UP000078492 UP000092444 UP000007755 UP000078540 UP000005205 UP000075809 UP000078541 UP000092443 UP000215335 UP000018467 UP000193380 UP000087266 UP000265040
Pfam
PF01412 ArfGap
Interpro
Gene 3D
ProteinModelPortal
H9JQR3
A0A2W1BUX4
A0A2A4K9L3
A0A2H1WVD6
A0A0N1PH52
A0A194Q1C5
+ More
A0A212ELU7 A0A2P8XE23 A0A1J1I3U8 A0A1W4URC5 A0A067QYD0 A0A2J7QGZ2 A0A1Y1KF72 B4N5G2 B4KYJ1 D6WF54 B4HC18 Q16YC8 A0A1S4FK08 Q29F15 A0A023EQB2 B4J3Q1 A0A0K8W3L1 A0A0A1XGT1 A0A182IQW6 A0A034W8H5 B4LDY5 A0A1I8QB38 A0A0M4F0T2 A0A084WHJ9 W8CBT7 Q9VNR9 A0A182QT18 A0A3B0JUA2 B4QLA0 B4IB37 A0A182WG71 N6TSB9 A0A182MYN4 B3MB80 B4PIP6 A0A182RTI9 A0A182MS58 A0A182P6I1 A0A0J7KIR1 Q5TN70 A0A182UH13 A0A182SV23 A0A182WZL7 A0A182V4A6 A0A182HQ63 A0A182JZ05 A0A1B0B6X3 A0A182Y1P5 A0A2M4BS69 A0A182FW68 A0A2M4AUR5 A0A0T6B6M6 A0A2M4AUX8 E9IUR0 E2ACB6 A0A0L0C8C7 A0A1B0EXZ7 A0A1I8MS43 B0WFZ5 A0A1A9WD82 A0A026WZM7 A0A3L8DPS2 A0A2M4BQU4 A0A0Q5UL26 A0A1A9VFA8 A0A1B0A2D3 W5JIQ5 A0A195DXY4 A0A1B0FB80 F4WZN9 A0A195B6X2 A0A158NH77 A0A1B6BW40 A0A151XCU8 A0A195FFI4 A0A1A9Y0Y5 A0A1B6ERF1 A0A1B6HEL0 A0A232FKF6 A0A3B1JWN3 A0A293MU96 A0A2M4AXM3 A0A1B6M7L5 A0A060X5C3 A0A1S3L5U4 A0A3Q1HIP3 A0A1A8IR40 A0A2R5LMS0
A0A212ELU7 A0A2P8XE23 A0A1J1I3U8 A0A1W4URC5 A0A067QYD0 A0A2J7QGZ2 A0A1Y1KF72 B4N5G2 B4KYJ1 D6WF54 B4HC18 Q16YC8 A0A1S4FK08 Q29F15 A0A023EQB2 B4J3Q1 A0A0K8W3L1 A0A0A1XGT1 A0A182IQW6 A0A034W8H5 B4LDY5 A0A1I8QB38 A0A0M4F0T2 A0A084WHJ9 W8CBT7 Q9VNR9 A0A182QT18 A0A3B0JUA2 B4QLA0 B4IB37 A0A182WG71 N6TSB9 A0A182MYN4 B3MB80 B4PIP6 A0A182RTI9 A0A182MS58 A0A182P6I1 A0A0J7KIR1 Q5TN70 A0A182UH13 A0A182SV23 A0A182WZL7 A0A182V4A6 A0A182HQ63 A0A182JZ05 A0A1B0B6X3 A0A182Y1P5 A0A2M4BS69 A0A182FW68 A0A2M4AUR5 A0A0T6B6M6 A0A2M4AUX8 E9IUR0 E2ACB6 A0A0L0C8C7 A0A1B0EXZ7 A0A1I8MS43 B0WFZ5 A0A1A9WD82 A0A026WZM7 A0A3L8DPS2 A0A2M4BQU4 A0A0Q5UL26 A0A1A9VFA8 A0A1B0A2D3 W5JIQ5 A0A195DXY4 A0A1B0FB80 F4WZN9 A0A195B6X2 A0A158NH77 A0A1B6BW40 A0A151XCU8 A0A195FFI4 A0A1A9Y0Y5 A0A1B6ERF1 A0A1B6HEL0 A0A232FKF6 A0A3B1JWN3 A0A293MU96 A0A2M4AXM3 A0A1B6M7L5 A0A060X5C3 A0A1S3L5U4 A0A3Q1HIP3 A0A1A8IR40 A0A2R5LMS0
Ontologies
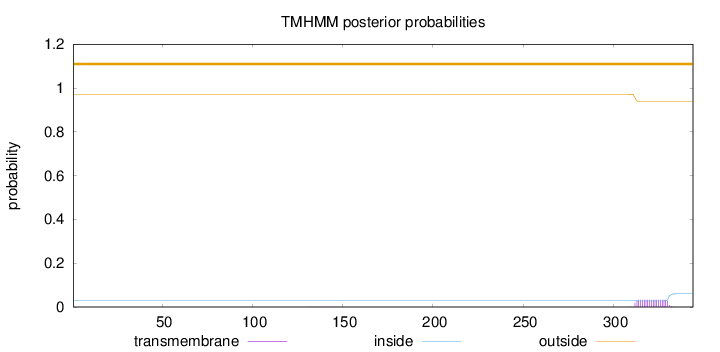
Topology
Length:
344
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.62451
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02858
outside
1 - 344
Population Genetic Test Statistics
Pi
26.715049
Theta
18.834948
Tajima's D
1.620735
CLR
0.454536
CSRT
0.821208939553022
Interpretation
Uncertain
